4WFS
 
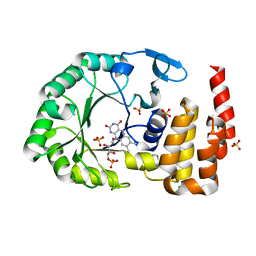 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
4WFT
 
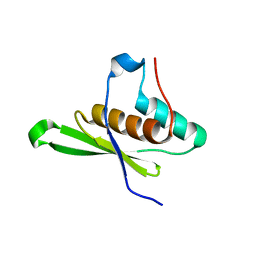 | | Crystal structure of tRNA-dihydrouridine(20) synthase dsRBD domain | | Descriptor: | tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
6EI9
 
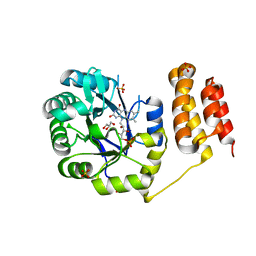 | | Crystal structure of E. coli tRNA-dihydrouridine synthase B (DusB) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Hamdane, D. | | Deposit date: | 2017-09-18 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Unveiling structural and functional divergences of bacterial tRNA dihydrouridine synthases: perspectives on the evolution scenario.
Nucleic Acids Res., 46, 2018
|
|
7TQB
 
 | |
6EI8
 
 | |
7TQA
 
 | | Crystal Structure of monoclonal S9.6 Fab | | Descriptor: | Fab S9.6 heavy chain, Fab S9.6 light chain, GLYCEROL, ... | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Structural basis of R-loop recognition by the S9.6 monoclonal antibody.
Nat Commun, 13, 2022
|
|
6EZB
 
 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain Q305K mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Bou-Nader, C, Bregeon, D, Pecqueur, L, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
6EZA
 
 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain E294K mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Bregeon, D, Pecqueur, L, Vincent, G, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
6EZC
 
 | | Crystal Structure of human tRNA-dihydrouridine(20) synthase catalytic domain E294K Q305K double mutant | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Bou-Nader, C, Bregeon, D, Vincent, G, Fontecave, M, Hamdane, D. | | Deposit date: | 2017-11-14 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Electrostatic Potential in the tRNA Binding Evolution of Dihydrouridine Synthases.
Biochemistry, 57, 2018
|
|
7NDZ
 
 | | ThyX reconstituted with N5-carbinolamine flavin | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
7NDW
 
 | | ThyX-FADH2 soaked with 20 mM Formaldehyde | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent thymidylate synthase, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2021-02-02 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An enzymatic activation of formaldehyde for nucleotide methylation.
Nat Commun, 12, 2021
|
|
7MRL
 
 | |
5OC5
 
 | |
5NL1
 
 | | Shigella IpaA-VBS3/TBS in complex with the Talin VBS1 domain 488-512 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Invasin IpaA, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Valencia-Gallardo, C, Fontecave, M, Tran Van Nhieu, G. | | Deposit date: | 2017-04-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Shigella IpaA Binding to Talin Stimulates Filopodial Capture and Cell Adhesion.
Cell Rep, 26, 2019
|
|
5OC4
 
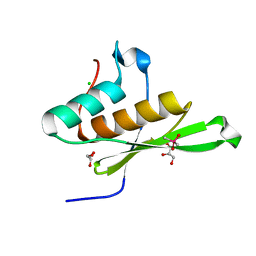 | | Crystal structure of human tRNA-dihydrouridine(20) synthase dsRBD R361A-R362A mutant | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bou-nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2017-06-29 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular basis for transfer RNA recognition by the double-stranded RNA-binding domain of human dihydrouridine synthase 2.
Nucleic Acids Res., 47, 2019
|
|
9C9R
 
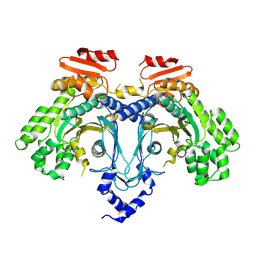 | |
9C9Q
 
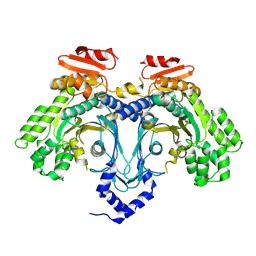 | |
5OC6
 
 | |
9DE7
 
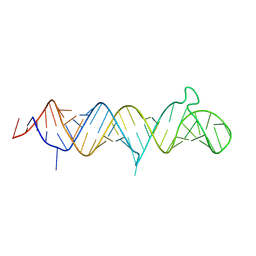 | | Structure of full-length HIV TAR RNA G16A/A17G | | Descriptor: | RNA (58-MER) | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of complete HIV-1 TAR RNA portray a dynamic platform poised for protein binding and structural remodeling.
Nat Commun, 16, 2025
|
|
9DE8
 
 | | Structure of full-length HIV TAR RNA soaked in CaCl2 | | Descriptor: | CALCIUM ION, RNA (56-MER) | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structures of complete HIV-1 TAR RNA portray a dynamic platform poised for protein binding and structural remodeling.
Nat Commun, 16, 2025
|
|
9DE6
 
 | | Structure of full-length HIV TAR RNA | | Descriptor: | MAGNESIUM ION, RNA (57-MER) | | Authors: | Bou-Nader, C, Zhang, J. | | Deposit date: | 2024-08-28 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of complete HIV-1 TAR RNA portray a dynamic platform poised for protein binding and structural remodeling.
Nat Commun, 16, 2025
|
|
9DE5
 
 | |
8FT9
 
 | | Crystal structure of a new antigen receptor variable domain from nurse sharks | | Descriptor: | GLYCEROL, PHOSPHATE ION, VNAR B6 | | Authors: | Yerabham, A.S.K, Hsieh, C.M, Fu, Y, Bou-Nader, C, Cachau, R, Zhang, J, Ho, M. | | Deposit date: | 2023-01-11 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of a new antigen receptor variable domain from nurse sharks
To Be Published
|
|
6OL3
 
 | | Crystal structure of an adenovirus virus-associated RNA | | Descriptor: | Adenovirus Virus-Associated (VA) RNA I apical and central domains, POTASSIUM ION | | Authors: | Hood, I.V, Gordon, J.M, Bou-Nader, C, Henderson, F.V, Bahmanjah, S, Zhang, J. | | Deposit date: | 2019-04-15 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Crystal structure of an adenovirus virus-associated RNA.
Nat Commun, 10, 2019
|
|
