7AA5
 
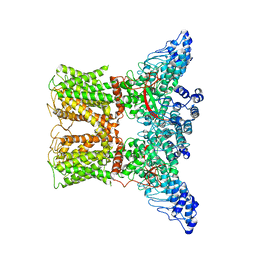 | | Human TRPV4 structure in presence of 4a-PDD | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 4,Green fluorescent protein | | Authors: | Botte, M, Ulrich, A.K.G, Adaixo, R, Gnutt, D, Brockmann, A, Bucher, D, Chami, M, Bocquet, M, Ebbinghaus-Kintscher, U, Puetter, V, Becker, A, Egner, U, Stahlberg, H, Hennig, M, Holton, S.J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Cryo-EM structural studies of the agonist complexed human TRPV4 ion-channel reveals novel structural rearrangements resulting in an open-conformation
To Be Published
|
|
7OMT
 
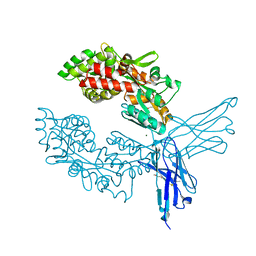 | | Crystal structure of ProMacrobody 21 with bound maltose | | Descriptor: | HEXAETHYLENE GLYCOL, MAGNESIUM ION, ProMacrobody 21, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7OMM
 
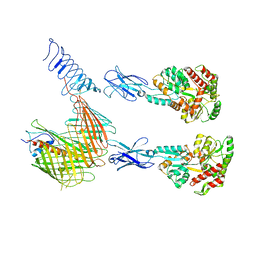 | | Cryo-EM structure of N. gonorhoeae LptDE in complex with ProMacrobodies (MBPs have not been built de novo) | | Descriptor: | LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ProMacrobody 21,Maltodextrin-binding protein, ... | | Authors: | Botte, M, Ni, D, Schenck, S, Zimmermann, I, Chami, M, Bocquet, N, Egloff, P, Bucher, D, Trabuco, M, Cheng, R.K.Y, Brunner, J.D, Seeger, M.A, Stahlberg, H, Hennig, M. | | Deposit date: | 2021-05-24 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of a LptDE transporter in complex with Pro-macrobodies offer insight into lipopolysaccharide translocation.
Nat Commun, 13, 2022
|
|
7PQT
 
 | | Apo human Kv3.1 cryo-EM structure | | Descriptor: | POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
7PQU
 
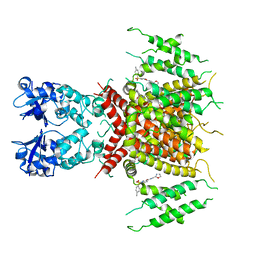 | | Ligand-bound human Kv3.1 cryo-EM structure (Lu AG00563) | | Descriptor: | 1-(4-methylphenyl)sulfonyl-N-(1,3-oxazol-2-ylmethyl)pyrrole-3-carboxamide, POTASSIUM ION, Potassium voltage-gated channel subfamily C member 1 | | Authors: | Botte, M, Huber, S, Bucher, D, Klint, J.K, Rodriguez, D, Tagmose, L, Chami, M, Cheng, R, Hennig, M, Abdul Rhaman, W. | | Deposit date: | 2021-09-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Apo and ligand-bound high resolution Cryo-EM structures of the human Kv3.1 channel reveal a novel binding site for positive modulators.
Pnas Nexus, 1, 2022
|
|
1H5P
 
 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
8OO6
 
 | | Pol I bound to extended and displaced DNA section - closed conformation | | Descriptor: | DNA polymerase I, Displaced primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OOY
 
 | | Pol I bound to extended and displaced DNA section - open conformation | | Descriptor: | DNA polymerase I, Displacing Primer, Extending Primer, ... | | Authors: | Botto, M, Borsellini, A, Lamers, M.H. | | Deposit date: | 2023-04-06 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A four-point molecular handover during Okazaki maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
1XWH
 
 | | NMR structure of the first phd finger of autoimmune regulator protein (AIRE1): insights into apeced | | Descriptor: | Autoimmune regulator, ZINC ION | | Authors: | Bottomley, M.J, Stier, G, Krasotkina, J, Legube, G, Simon, B, Akhtar, A, Sattler, M, Musco, G. | | Deposit date: | 2004-11-01 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease
J.Biol.Chem., 280, 2005
|
|
2W2O
 
 | | PCSK9-deltaC D374Y mutant bound to WT EGF-A of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2W2Q
 
 | | PCSK9-deltaC D374H mutant bound to WT EGF-A of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2W2P
 
 | | PCSK9-deltaC D374A mutant bound to WT EGF-A of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2W2M
 
 | | WT PCSK9-DELTAC BOUND TO WT EGF-A OF LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2W2N
 
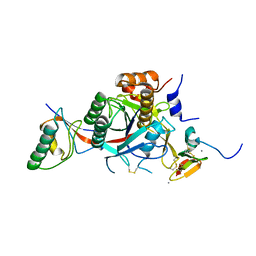 | | WT PCSK9-deltaC bound to EGF-A H306Y mutant of LDLR | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, PROPROTEIN CONVERTASE SUBTILISIN/KEXIN TYPE 9 | | Authors: | Bottomley, M.J, Cirillo, A, Orsatti, L, Ruggeri, L, Fisher, T.S, Santoro, J.C, Cummings, R.T, Cubbon, R.M, Lo Surdo, P, Calzetta, A, Noto, A, Baysarowich, J, Mattu, M, Talamo, F, De Francesco, R, Sparrow, C.P, Sitlani, A, Carfi, A. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
J.Biol.Chem., 284, 2009
|
|
2VQO
 
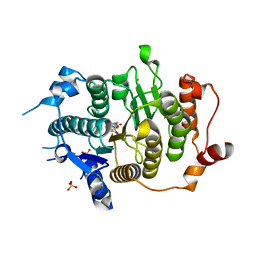 | | Structure of HDAC4 catalytic domain with a gain-of-function muation bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQQ
 
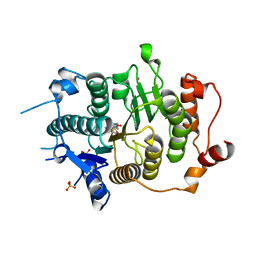 | | Structure of HDAC4 catalytic domain (a double cysteine-to-alanine mutant) bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQV
 
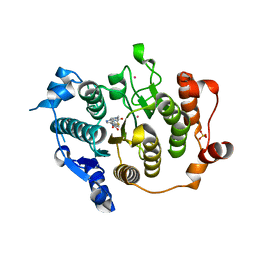 | | Structure of HDAC4 catalytic domain with a gain-of-function mutation bound to a hydroxamic acid inhibitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2UV0
 
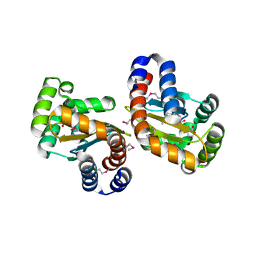 | | Structure of the P. aeruginosa LasR ligand-binding domain bound to its autoinducer | | Descriptor: | N-3-OXO-DODECANOYL-L-HOMOSERINE LACTONE, TRANSCRIPTIONAL ACTIVATOR PROTEIN LASR | | Authors: | Bottomley, M.J, Muraglia, E, Bazzo, R, Carfi, A. | | Deposit date: | 2007-03-08 | | Release date: | 2007-03-27 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Insights Into Quorum Sensing in the Human Pathogen Pseudomonas Aeruginosa from the Structure of the Virulence Regulator Lasr Bound to its Autoinducer.
J.Biol.Chem., 282, 2007
|
|
2VQW
 
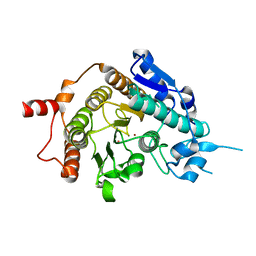 | | Structure of inhibitor-free HDAC4 catalytic domain (with gain-of- function mutation His332Tyr) | | Descriptor: | HISTONE DEACETYLASE 4, POTASSIUM ION, ZINC ION | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQM
 
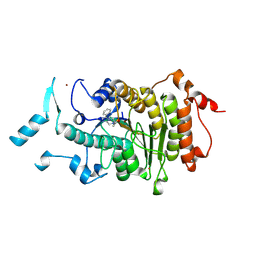 | | Structure of HDAC4 catalytic domain bound to a hydroxamic acid inhbitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQJ
 
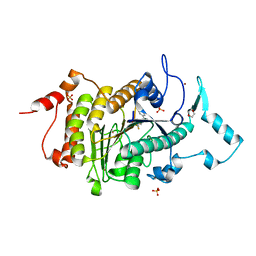 | | Structure of HDAC4 catalytic domain bound to a trifluoromethylketone inhbitor | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
7Q21
 
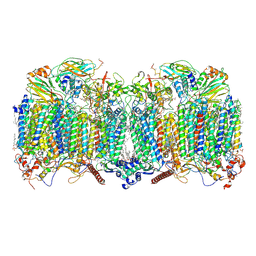 | | III2-IV2 respiratory supercomplex from Corynebacterium glutamicum | | Descriptor: | (1R)-2-(dodecanoyloxy)-1-[(phosphonooxy)methyl]ethyl tetradecanoate, (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Kovalova, T, Moe, A, Krol, S, Yanofsky, D.J, Bott, M, Sjostrand, D, Rubinstein, J.L, Hogbom, M, Brzezinski, P. | | Deposit date: | 2021-10-22 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The respiratory supercomplex from C. glutamicum.
Structure, 30, 2022
|
|
3F51
 
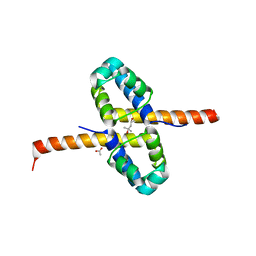 | | Crystal Structure of the clp gene regulator ClgR from Corynebacterium glutamicum | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
4QCJ
 
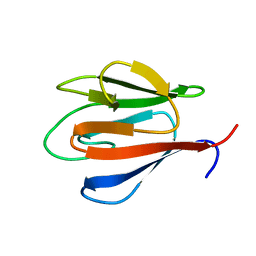 | | Crystal Structure of OdhI from Corynebacterium glutamicum | | Descriptor: | Oxoglutarate dehydrogenase inhibitor | | Authors: | Labahn, J, Raasch, K, Eggeling, L, Bocola, M, Leitner, A, Bott, M. | | Deposit date: | 2014-05-12 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction of 2-oxoglutarate dehydrogenase OdhA with its inhibitor OdhI in Corynebacterium glutamicum: Mutants and a model.
J.Biotechnol., 191, 2014
|
|
3F52
 
 | | Crystal structure of the clp gene regulator ClgR from C. glutamicum | | Descriptor: | GLYCEROL, clp gene regulator (ClgR) | | Authors: | Russo, S, Schweitzer, J.E, Polen, T, Bott, M, Pohl, E. | | Deposit date: | 2008-11-03 | | Release date: | 2008-11-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the caseinolytic protease gene regulator, a transcriptional activator in actinomycetes
J.Biol.Chem., 284, 2009
|
|
