6NN6
 
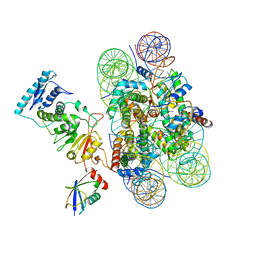 | | Structure of Dot1L-H2BK120ub nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Anderson, C.J, Baird, M.R, Hsu, A, Barbour, E.H, Koyama, Y, Borgnia, M.J, McGinty, R.K. | | Deposit date: | 2019-01-14 | | Release date: | 2019-02-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase.
Cell Rep, 26, 2019
|
|
6NR3
 
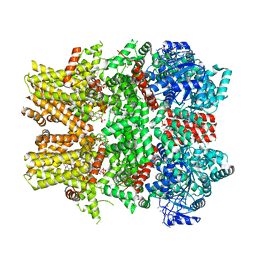 | | Cryo-EM structure of the TRPM8 ion channel in complex with high occupancy icilin, PI(4,5)P2, and calcium | | Descriptor: | (2S)-1-{[(R)-hydroxy{[(1R,2R,3S,4R,5R,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl icosa-5,8,11,14-tetraenoate, CALCIUM ION, Icilin, ... | | Authors: | Yin, Y, Le, S.C, Hsu, A.L, Borgnia, M.J, Yang, H, Lee, S.-Y. | | Deposit date: | 2019-01-22 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of cooling agent and lipid sensing by the cold-activated TRPM8 channel.
Science, 363, 2019
|
|
6OF4
 
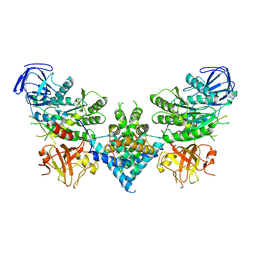 | | Precursor ribosomal RNA processing complex, apo-state. | | Descriptor: | CLP1_P domain-containing protein, Ribonuclease | | Authors: | Pillon, M.C, Hsu, A.L, Krahn, J.M, Williams, J.G, Goslen, K.H, Sobhany, M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2019-03-28 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM reveals active site coordination within a multienzyme pre-rRNA processing complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6HMB
 
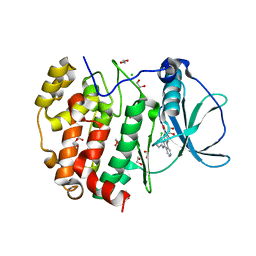 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 Gene product) IN COMPLEX WITH the inhibitor CX-4945 (Silmitasertib) | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3-chlorophenyl)amino]benzo[c][2,6]naphthyridine-8-carboxylic acid, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6HMQ
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 GENE PRODUCT) IN COMPLEX WITH THE BENZOTRIAZOLE-TYPE INHIBITOR MB002 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(4,5,6,7-tetrabromo-1H-benzotriazol-1-yl)propan-1-ol, ... | | Authors: | Niefind, K, Lindenblatt, D, Applegate, V.M, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6HBN
 
 | | HIGH-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA/CSKN2A1 GENE PRODUCT) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR THN27 | | Descriptor: | 5-propan-2-yl-4-prop-2-enoxy-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Niefind, K, Hochscherf, J, Dimper, V, Witulski, B, Lindenblatt, D, Jose, J, Le Borgne, M. | | Deposit date: | 2018-08-10 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6HMD
 
 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA'; CSNK2A2 gene product) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR AR18 | | Descriptor: | 1,2-ETHANEDIOL, 5-[2-(diethylamino)ethyl]-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Niefind, K, Lindenblatt, D, Jose, J, Le Borgne, M. | | Deposit date: | 2018-09-12 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Diacritic Binding of an Indenoindole Inhibitor by CK2 alpha Paralogs Explored by a Reliable Path to Atomic Resolution CK2 alpha ' Structures.
Acs Omega, 4, 2019
|
|
6PKW
 
 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C2 symmetry (pseudo C4 symmetry) | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6PKV
 
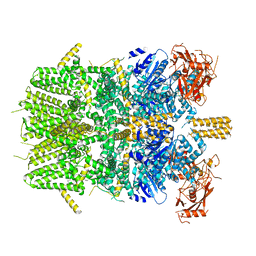 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C4 symmetry | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
8V55
 
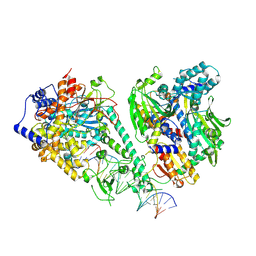 | | Human mitochondrial DNA polymerase gamma bound to a replication fork in an open conformation | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, M.J, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
6PKX
 
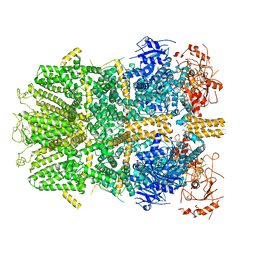 | | Cryo-EM structure of the zebrafish TRPM2 channel in the presence of ADPR and Ca2+ | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
8VIW
 
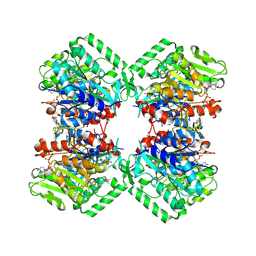 | | Cryo-EM structure of heparosan synthase 2 from Pasteurella multocida with polysaccharide in the GlcNAc-T active site | | Descriptor: | Heparosan synthase B, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Krahn, J.M, Pedersen, L.C, Liu, J, Stancanelli, E, Borgnia, M, Vivarette, E. | | Deposit date: | 2024-01-05 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and Functional Analysis of Heparosan Synthase 2 from Pasteurella multocida to Improve the Synthesis of Heparin
Acs Catalysis, 14, 2024
|
|
6UKP
 
 | | Apo mBcs1 | | Descriptor: | Mitochondrial chaperone BCS1 | | Authors: | Tang, W.K, Borgnia, M.J, Hsu, A.L, Xia, D. | | Deposit date: | 2019-10-05 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8DTI
 
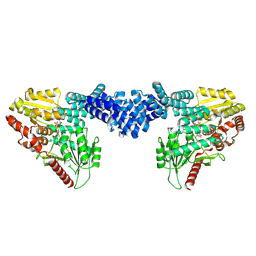 | | Cryo-EM structure of Arabidopsis SPY in complex with GDP-fucose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-BETA-L-FUCOPYRANOSE, Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTH
 
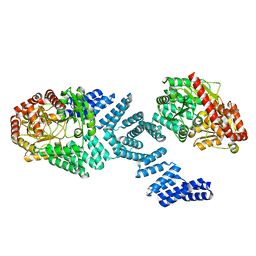 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 2 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTG
 
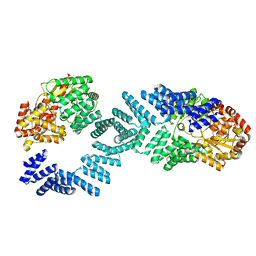 | | Cryo-EM structure of Arabidopsis SPY alternative conformation 1 | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
8DTF
 
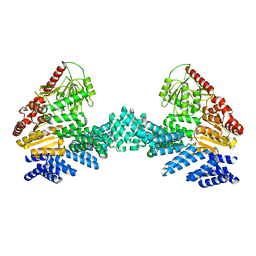 | | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs | | Descriptor: | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY | | Authors: | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M.J, Bartesaghi, A, Zhou, P. | | Deposit date: | 2022-07-25 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the full length Arabidopsis SPY with complete TPRs
Nat Commun, 2023
|
|
4CC8
 
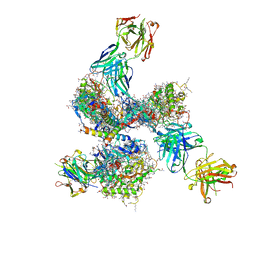 | | Pre-fusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy | | Descriptor: | GP120, GP41, MONOCLONAL ANTIBODY VRC03 FAB HEAVY CHAIN, ... | | Authors: | Bartesaghi, A, Merk, A, Borgnia, M.J, Milne, J.L.S, Subramaniam, S. | | Deposit date: | 2013-10-18 | | Release date: | 2013-10-30 | | Last modified: | 2018-01-10 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Prefusion Structure of Trimeric HIV-1 Envelope Glycoprotein Determined by Cryo-Electron Microscopy.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6PQP
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist BITC | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-benzylthioformamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PQQ
 
 | | Cryo-EM structure of human TRPA1 C621S mutant in the apo state | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily A member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PQO
 
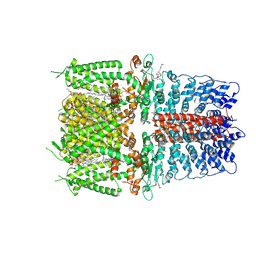 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist JT010 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-N-(3-methoxypropyl)acetamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
3EE6
 
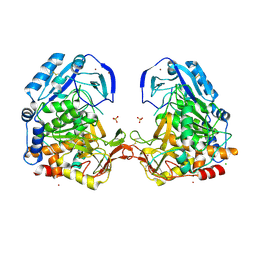 | | Crystal Structure Analysis of Tripeptidyl peptidase -I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Pal, A, Kraetzner, R, Grapp, M, Gruene, T, Schreiber, K, Granborg, M, Urlaub, H, Asif, A.R, Becker, S, Gartner, J, Sheldrick, G.M, Steinfeld, R. | | Deposit date: | 2008-09-04 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis
J.Biol.Chem., 284, 2009
|
|
5T7K
 
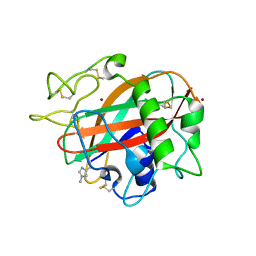 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
6OO7
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in nanodiscs | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OO5
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 4.2 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
