7NP0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (4-nitrophenyl)boronic acid. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION, p-nitrophenylboronic acid | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NM0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(2,6-dihydroxyphenyl)ethan-1-one. | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(oxidanyl)phenyl]ethanone, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNI
 
 | |
7NOR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with 2-fluoro-4-hydroxybenzonitrile. | | Descriptor: | 2-fluoro-4-hydroxybenzonitrile, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-(5-methoxy-1H-indol-3-yl)acetonitrile | | Descriptor: | 2-(5-methoxy-1~{H}-indol-3-yl)ethanenitrile, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NOV
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with (4-methyl-3-nitrophenyl)boronic acid. | | Descriptor: | (4-methyl-3-nitro-phenyl)-oxidanyl-oxidanylidene-boron, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLF
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in apo form. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLU
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(1H-indol-3-yl)ethan-1-one | | Descriptor: | 1-(1~{H}-indol-3-yl)ethanone, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.235 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN7
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with dimethyl 5-hydroxyisophthalate. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNW
 
 | | Crystal structure of Mycobacterium tuberculosis ArgF in complex with methyl 4-hydroxy-3-iodobenzoate. | | Descriptor: | Ornithine carbamoyltransferase, PHOSPHATE ION, methyl 3-iodanyl-4-oxidanyl-benzoate | | Authors: | Mendes, V, Gupta, P, Burgess, A, Sebastian-Perez, V, Cattermole, E, Meghir, C, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NFC
 
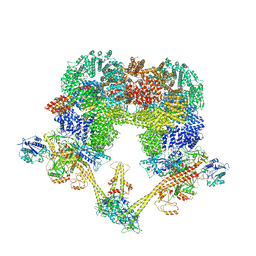 | | Cryo-EM structure of NHEJ super-complex (dimer) | | Descriptor: | DNA (27-MER), DNA (28-MER), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
7NFE
 
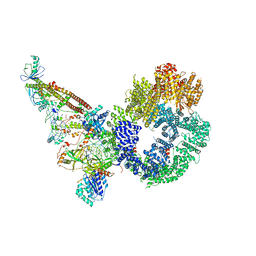 | | Cryo-EM structure of NHEJ super-complex (monomer) | | Descriptor: | DNA (5'-D(P*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*CP*TP*AP*TP*TP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(P*TP*AP*AP*TP*AP*AP*TP*AP*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*AP*G)-3'), DNA ligase 4, ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Kefala Stavridi, A, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2021-02-06 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Cryo-EM of NHEJ supercomplexes provides insights into DNA repair.
Mol.Cell, 81, 2021
|
|
1XES
 
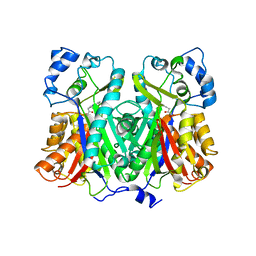 | | Crystal structure of stilbene synthase from Pinus sylvestris | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Dihydropinosylvin synthase | | Authors: | Ng, S.H, Chirgadze, D, Spiteller, D, Li, T.L, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2004-09-12 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of stilbene synthase from Pinus sylvestris
To be Published
|
|
1XET
 
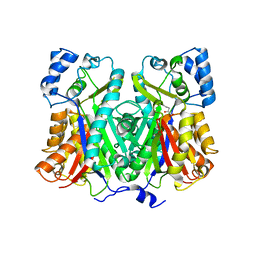 | | Crystal structure of stilbene synthase from Pinus sylvestris, complexed with methylmalonyl CoA | | Descriptor: | 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Dihydropinosylvin synthase, METHYLMALONYL-COENZYME A | | Authors: | Ng, S.H, Chirgadze, D, Spiteller, D, Li, T.L, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2004-09-12 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of stilbene synthase from Pinus sylvestris, complexed with methylmalonyl CoA
To be Published
|
|
3PHV
 
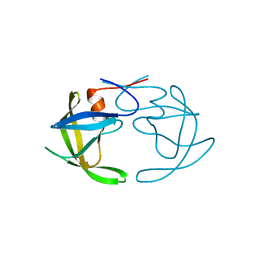 | | X-RAY ANALYSIS OF HIV-1 PROTEINASE AT 2.7 ANGSTROMS RESOLUTION CONFIRMS STRUCTURAL HOMOLOGY AMONG RETROVIRAL ENZYMES | | Descriptor: | UNLIGANDED HIV-1 PROTEASE | | Authors: | Lapatto, R, Blundell, T.L, Hemmings, A, Wilderspin, A, Wood, S.P, Danley, D.E, Geoghegan, K.F, Hawrylik, S.J, Hobart, P.M. | | Deposit date: | 1991-11-04 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes.
Nature, 342, 1989
|
|
1EPM
 
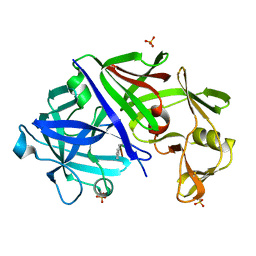 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, PS2, THR-PHE-GLN-ALA-PSA-LEU-ARG-GLU, ... | | Authors: | Crawford, M, Cooper, J.B, Strop, P, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1EPP
 
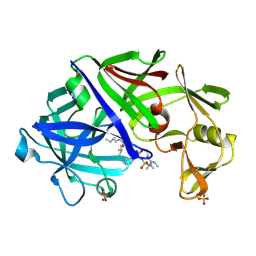 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EPN
 
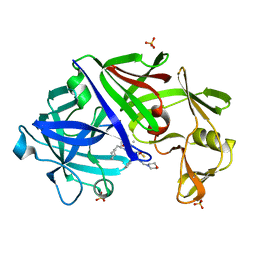 | | A STRUCTURAL COMPARISON OF 21 INHIBITOR COMPLEXES OF THE ASPARTIC PROTEINASE FROM ENDOTHIA PARASITICA | | Descriptor: | ENDOTHIAPEPSIN, N-(morpholin-4-ylcarbonyl)-L-phenylalanyl-N-[(1R,2S)-1-(cyclohexylmethyl)-2-hydroxy-3-(1-methylethoxy)-3-oxopropyl]-S-methyl-L-cysteinamide, SULFATE ION | | Authors: | Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
6MJY
 
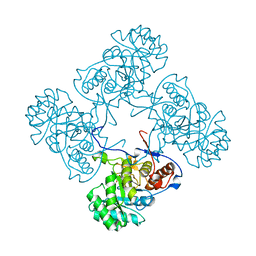 | |
6NVR
 
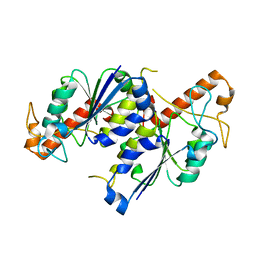 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in Apo form | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V.M, Blundell, T.L. | | Deposit date: | 2019-02-05 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.562 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6NW7
 
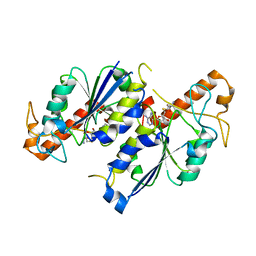 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with S-Adenosyl-L-homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
6NW6
 
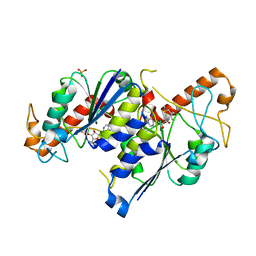 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with S-Adenosyl methionine | | Descriptor: | S-ADENOSYLMETHIONINE, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
8C15
 
 | | Aurora A kinase in complex with TPX2-inhibitor 3 | | Descriptor: | (1~{R},2~{R})-cyclohexane-1,2-dicarboxylic acid, 4-(4-chlorophenyl)-1~{H}-indole-6-carboxylic acid, ACETATE ION, ... | | Authors: | Fischer, G, Rocaboy, M, Blaszczyk, B, Moschetti, T, Wang, X, Scott, D.E, Coyne, A.G, Dagostin, C, Rooney, T, Bayly, A, Feng, J, Asteian, A, Alcaide-Lopez, A, Stockwell, S, Skidmore, J, Venkitaraman, A.R, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2022-12-20 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Selective inhibitors of the Aurora A-TPX2 protein-protein interaction exhibit in vivo efficacy as targeted anti-mitotic agents
To be published
|
|
1A0R
 
 | | HETEROTRIMERIC COMPLEX OF PHOSDUCIN/TRANSDUCIN BETA-GAMMA | | Descriptor: | FARNESYL, PHOSDUCIN, TRANSDUCIN (BETA SUBUNIT), ... | | Authors: | Loew, A, Ho, Y.-K, Blundell, T.L, Bax, B. | | Deposit date: | 1997-12-05 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phosducin induces a structural change in transducin beta gamma.
Structure, 6, 1998
|
|
6QQX
 
 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-[1-(pyridin-2-ylmethyl)indol-6-yl]-1~{H}-pyrazol-3-amine, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Fragment-based discovery of a new class of inhibitors targeting mycobacterial tRNA modification.
Nucleic Acids Res., 48, 2020
|
|
