6QRG
 
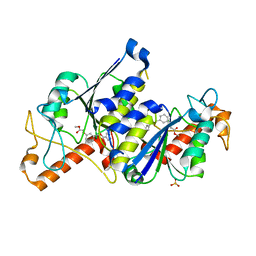 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | SULFATE ION, [3-(2-methoxy-2-oxidanylidene-ethoxy)phenyl]methyl 1~{H}-pyrazole-4-carboxylate, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
3V42
 
 | |
6SQL
 
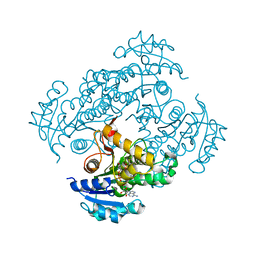 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and N-(3-(aminomethyl)phenyl)-5-chloro-3-methylbenzo[b]thiophene-2-sulfonamide | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ~{N}-[3-(aminomethyl)phenyl]-5-chloranyl-3-methyl-1-benzothiophene-2-sulfonamide | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-04 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQB
 
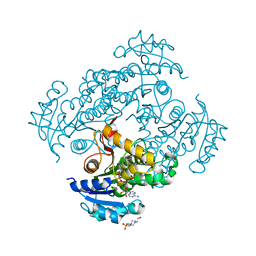 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-(3-chlorophenyl)propanoic acid | | Descriptor: | 3-(3-chlorophenyl)propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.774 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQD
 
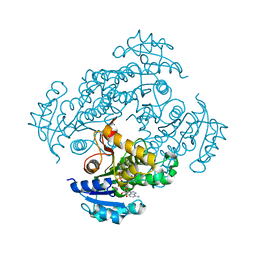 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 2-pyrazol-1-ylbenzoic acid | | Descriptor: | 2-pyrazol-1-ylbenzoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6TH2
 
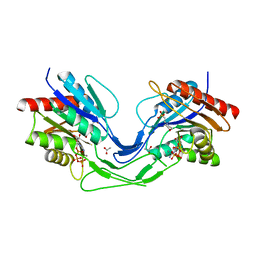 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
3W1G
 
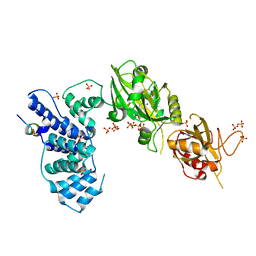 | | Crystal Structure of Human DNA ligase IV-Artemis Complex (Native) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Artemis-derived peptide, DNA ligase 4, ... | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2012-11-15 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Structure, 21, 2013
|
|
3W03
 
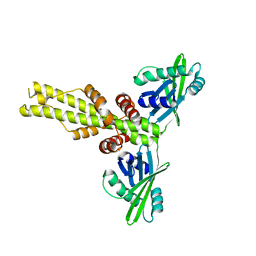 | | XLF-XRCC4 complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Wu, Q, Ochi, T, Matak-Vinkovic, D, Robinson, C.V, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2012-10-17 | | Release date: | 2012-11-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (8.492 Å) | | Cite: | Non-homologous end-joining partners in a helical dance: structural studies of XLF-XRCC4 interactions
Biochem.Soc.Trans., 39, 2011
|
|
6TGV
 
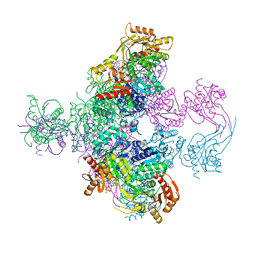 | | Crystal structure of Mycobacterium smegmatis CoaBC in complex with CTP and FMN | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein CoaBC, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
6SQ9
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-hydroxynaphthalene-2-carboxylic acid | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
3W1B
 
 | |
6SQ7
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 2-(4-chloro-3-nitrobenzoyl)benzoic acid | | Descriptor: | 2-(4-chloranyl-3-nitro-phenyl)carbonylbenzoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
6SQ5
 
 | | Crystal structure of M. tuberculosis InhA in complex with NAD+ and 3-[3-(trifluoromethyl)phenyl]prop-2-enoic acid | | Descriptor: | (~{E})-3-[3-(trifluoromethyl)phenyl]prop-2-enoic acid, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Mendes, V, Sabbah, M, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Fragment-Based Design ofMycobacterium tuberculosisInhA Inhibitors.
J.Med.Chem., 63, 2020
|
|
3VNN
 
 | | Crystal Structure of a sub-domain of the nucleotidyltransferase (adenylation) domain of human DNA ligase IV | | Descriptor: | DNA ligase 4 | | Authors: | Ochi, T, Wu, Q, Chirgadze, D.Y, Grossmann, J.G, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2012-01-17 | | Release date: | 2012-06-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structural insights into the role of domain flexibility in human DNA ligase IV
Structure, 20, 2012
|
|
6THC
 
 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP and (4-hydroxyphenyl)(2,3,4-trihydroxyphenyl)methanone | | Descriptor: | (4-hydroxyphenyl)-[2,3,4-tris(oxidanyl)phenyl]methanone, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
3W5O
 
 | | Crystal Structure of Human DNA ligase IV | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA ligase 4, SULFATE ION | | Authors: | Gu, X, Ochi, T, Blundell, T.L. | | Deposit date: | 2013-02-02 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structure of the catalytic region of DNA ligase IV in complex with an artemis fragment sheds light on double-strand break repair
Structure, 21, 2013
|
|
3WTD
 
 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
3TM7
 
 | | Processed Aspartate Decarboxylase Mutant with Asn72 mutated to Ala | | Descriptor: | Aspartate 1-decarboxylase alpha chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Webb, M.E, Lobley, C.M.C, Soliman, F, Kilkenny, M.L, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2011-08-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Escherichia coli aspartate alpha-decarboxylase Asn72Ala: probing the role of Asn72 in pyruvoyl cofactor formation
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WTF
 
 | | Structure of PAXX | | Descriptor: | Uncharacterized protein C9orf142 | | Authors: | Ochi, T, Blundell, T.L. | | Deposit date: | 2014-04-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.451 Å) | | Cite: | DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair.
Science, 347, 2015
|
|
5J1R
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with 3-(furan-3-yl)-1-(pyrrolidin-1-yl)propan-1-one at 1.92A resolution | | Descriptor: | 3-(furan-3-yl)-1-(pyrrolidin-1-yl)propan-1-one, EthR | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
5J5R
 
 | | M. thermoresistible GuaB2 delta-CBS in complex with inhibitor VCC234718 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, cyclohexyl{4-[(isoquinolin-5-yl)sulfonyl]piperazin-1-yl}methanone | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-04-03 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Inosine Monophosphate Dehydrogenase, GuaB2, Is a Vulnerable New Bactericidal Drug Target for Tuberculosis.
ACS Infect Dis, 3, 2017
|
|
5J4L
 
 | | Apo-structure of humanised RadA-mutant humRadA22F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JIJ
 
 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase (APO form). | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-04-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
5JEE
 
 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5K42
 
 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a complex with GDP-glucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Alpha,alpha-trehalose-phosphate synthase, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-20 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
