5LB4
 
 | | Apo-structure of humanised RadA-mutant humRadA14 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LBI
 
 | | Apo-structure of humanised RadA-mutant humRadA3 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5L3K
 
 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a ternary complex with ADP and fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
4D6P
 
 | | RADA C-TERMINAL ATPASE DOMAIN FROM PYROCOCCUS FURIOSUS BOUND TO AMPPNP | | Descriptor: | DNA REPAIR AND RECOMBINATION PROTEIN RADA, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Marsh, M.E, Ehebauer, M.T, Scott, D, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2014-11-13 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | ATP Half-Sites in Rada and Rad51 Recombinases Bind Nucleotides
FEBS Open Bio, 6, 2016
|
|
4KL9
 
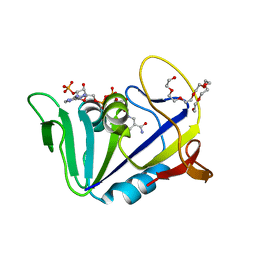 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in the space group C2 | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KIJ
 
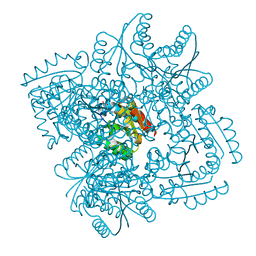 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinase dehydratase from Mycobacterium tuberculosis - compound 35c [3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid] | | Descriptor: | 3,4-dihydroxy-5-(3-nitrophenoxy)benzoic acid, 3-dehydroquinate dehydratase, CHLORIDE ION | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KIW
 
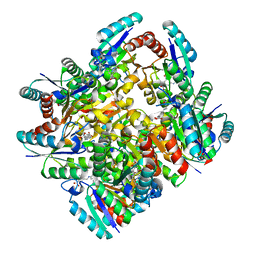 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49e [5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)amino]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4KG0
 
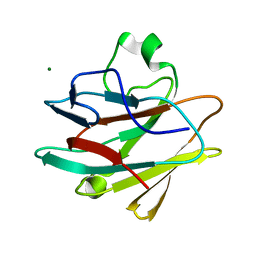 | | Crystal structure of the drosophila melanogaster neuralized-nhr1 domain | | Descriptor: | MAGNESIUM ION, Protein neuralized | | Authors: | Gupta, D, Ehebauer, M.T, Chirgadze, D.Y, Bolanos-Garcia, V.M, Blundell, T.L. | | Deposit date: | 2013-04-28 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure, biochemical and biophysical characterisation of NHR1 domain of E3 Ubiquitin ligase neutralized
Advances in Enzyme Research, 1, 2013
|
|
4KM2
 
 | |
4H7X
 
 | |
4KM0
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with pyrimethamine | | Descriptor: | 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-07 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KLX
 
 | |
4CJM
 
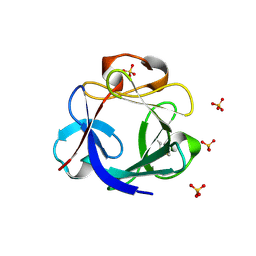 | | Crystal structure of human FGF18 | | Descriptor: | FIBROBLAST GROWTH FACTOR 18, SULFATE ION | | Authors: | Brown, A, Adam, L.E, Blundell, T.L. | | Deposit date: | 2013-12-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Crystal Structure of Fibroblast Growth Factor 18 (Fgf18)
Protein Cell, 5, 2014
|
|
4KNE
 
 | | Crystal structure of dihydrofolate reductase from Mycobacterium tuberculosis in complex with cycloguanil | | Descriptor: | 1-(4-chlorophenyl)-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Dihydrofolate reductase | | Authors: | Dias, M.V.B, Tyrakis, P, Blundell, T.L. | | Deposit date: | 2013-05-09 | | Release date: | 2013-12-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mycobacterium tuberculosis Dihydrofolate Reductase Reveals Two Conformational States and a Possible Low Affinity Mechanism to Antifolate Drugs.
Structure, 22, 2014
|
|
4KIU
 
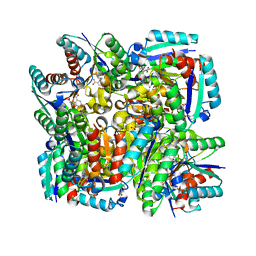 | | Design and structural analysis of aromatic inhibitors of type II dehydroquinate dehydratase from Mycobacterium tuberculosis - compound 49d [5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid] | | Descriptor: | 3-dehydroquinate dehydratase, 5-[(3-nitrobenzyl)oxy]benzene-1,3-dicarboxylic acid | | Authors: | Dias, M.V.B, Howard, N.G, Blundell, T.L, Abell, C. | | Deposit date: | 2013-05-02 | | Release date: | 2014-05-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Structural Analysis of Aromatic Inhibitors of Type II Dehydroquinase from Mycobacterium tuberculosis.
Chemmedchem, 10, 2015
|
|
4G5Y
 
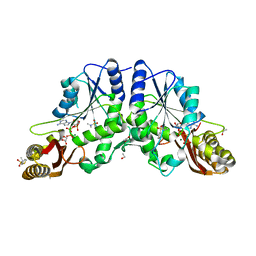 | | Crystal Structure of Mycobacterium tuberculosis Pantothenate synthetase in a ternary complex with ATP and N,N-DIMETHYLTHIOPHENE-3-SULFONAMIDE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Silvestre, H.L, Blundell, T.L, Abell, C. | | Deposit date: | 2012-07-18 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Integrated biophysical approach to fragment screening and validation for fragment-based lead discovery.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1IK9
 
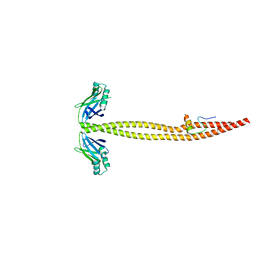 | | CRYSTAL STRUCTURE OF A XRCC4-DNA LIGASE IV COMPLEX | | Descriptor: | DNA LIGASE IV, DNA REPAIR PROTEIN XRCC4 | | Authors: | Sibanda, B.L, Critchlow, S.E, Begun, J, Pei, X.Y, Jackson, S.P, Blundell, T.L, Pellegrini, L. | | Deposit date: | 2001-05-03 | | Release date: | 2001-11-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of an Xrcc4-DNA ligase IV complex.
Nat.Struct.Biol., 8, 2001
|
|
1IHO
 
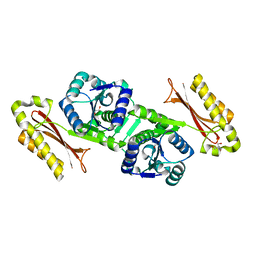 | | CRYSTAL APO-STRUCTURE OF PANTOTHENATE SYNTHETASE FROM E. COLI | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PANTOATE--BETA-ALANINE LIGASE | | Authors: | von Delft, F, Lewendon, A, Dhanaraj, V, Blundell, T.L, Abell, C, Smith, A. | | Deposit date: | 2001-04-19 | | Release date: | 2001-05-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of E. coli pantothenate synthetase confirms it as a member of the cytidylyltransferase superfamily.
Structure, 9, 2001
|
|
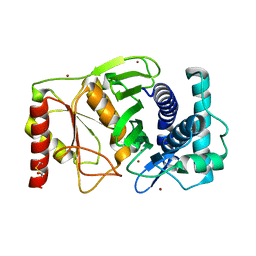
1K9Z
 
 | |
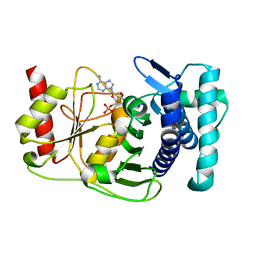
1KA0
 
 | |
1KS9
 
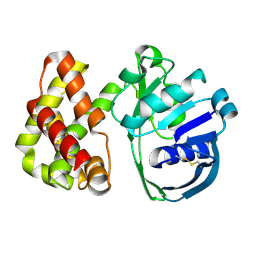 | | Ketopantoate Reductase from Escherichia coli | | Descriptor: | 2-DEHYDROPANTOATE 2-REDUCTASE | | Authors: | Matak-Vinkovic, D, Vinkovic, M, Saldanha, S.A, Ashurst, J.A, von Delft, F, Inoue, T, Miguel, R.N, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism.
Biochemistry, 40, 2001
|
|
1JP4
 
 | | Crystal Structure of an Enzyme Displaying both Inositol-Polyphosphate 1-Phosphatase and 3'-Phosphoadenosine-5'-Phosphate Phosphatase Activities | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, ... | | Authors: | Patel, S, Yenush, L, Rodriguez, P.L, Serrano, R, Blundell, T.L. | | Deposit date: | 2001-08-01 | | Release date: | 2001-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal structure of an enzyme displaying both inositol-polyphosphate-1-phosphatase and 3'-phosphoadenosine-5'-phosphate phosphatase activities: a novel target of lithium therapy.
J.Mol.Biol., 315, 2002
|
|
1K9Y
 
 | | The PAPase Hal2p complexed with magnesium ions and reaction products: AMP and inorganic phosphate | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-MERCAPTOETHANOL, Halotolerance protein HAL2, ... | | Authors: | Patel, S, Albert, A, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural enzymology of Li(+)-sensitive/Mg(2+)-dependent phosphatases.
J.Mol.Biol., 320, 2002
|
|
1KA1
 
 | | The PAPase Hal2p complexed with calcium and magnesium ions and reaction substrate: PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CALCIUM ION, ... | | Authors: | Patel, S, Albert, A, Blundell, T.L. | | Deposit date: | 2001-10-31 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural enzymology of Li(+)-sensitive/Mg(2+)-dependent phosphatases.
J.Mol.Biol., 320, 2002
|
|
1M3U
 
 | | Crystal Structure of Ketopantoate Hydroxymethyltransferase complexed the Product Ketopantoate | | Descriptor: | 3-methyl-2-oxobutanoate hydroxymethyltransferase, KETOPANTOATE, MAGNESIUM ION | | Authors: | von Delft, F, Inoue, T, Saldanha, S.A, Ottenhof, H.H, Dhanaraj, V, Witty, M, Abell, C, Smith, A.G, Blundell, T.L. | | Deposit date: | 2002-06-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E. coli Ketopantoate Hydroxymethyl Transferase Complexed with Ketopantoate and Mg(2+), Solved by Locating 160 Selenomethionine Sites.
Structure, 11, 2003
|
|
