3T92
 
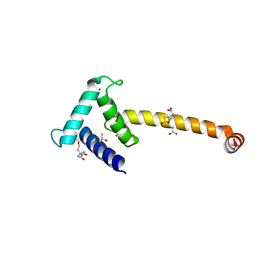 | | Crystal structure of the Taz2:C/EBPepsilon-TAD chimera protein | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, ACETONE, HISTONE ACETYLTRANSFERASE P300 TAZ2-CCAAT/ENHANCER-BINDING PROTEIN EPSILON, ... | | Authors: | Bhaumik, P, Maria, M. | | Deposit date: | 2011-08-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into interactions of C/EBP transcriptional activators with the Taz2 domain of p300.
Acta Crystallogr. D Biol. Crystallogr., 70, 2014
|
|
3FNS
 
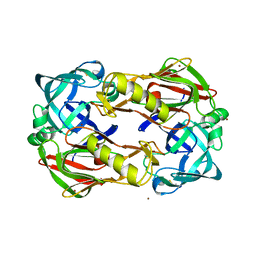 | |
3FNT
 
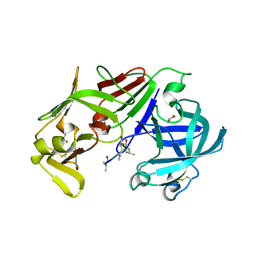 | | Crystal structure of pepstatin A bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | 1,2-ETHANEDIOL, HAP protein, Inhibitor, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
3FNU
 
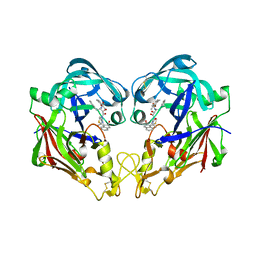 | | Crystal structure of KNI-10006 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, HAP protein | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2008-12-26 | | Release date: | 2009-05-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the histo-aspartic protease (HAP) from Plasmodium falciparum.
J.Mol.Biol., 388, 2009
|
|
1TJ7
 
 | | Structure determination and refinement at 2.44 A resolution of Argininosuccinate lyase from E. coli | | Descriptor: | Argininosuccinate lyase, GLYCEROL, PHOSPHATE ION | | Authors: | Bhaumik, P, Koski, M.K, Bergman, U, Wierenga, R.K. | | Deposit date: | 2004-06-03 | | Release date: | 2004-10-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure determination and refinement at 2.44 A resolution of argininosuccinate lyase from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3QRV
 
 | |
3QVC
 
 | |
3QVI
 
 | | Crystal structure of KNI-10395 bound histo-aspartic protease (HAP) from Plasmodium falciparum | | Descriptor: | (4R)-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-3-[(2S,3S)-2-hydroxy-3-{[S-methyl-N-(phenylacetyl)-L-cysteinyl]ami no}-4-phenylbutanoyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-25 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation and inhibition of histo-aspartic protease from Plasmodium falciparum.
Biochemistry, 50, 2011
|
|
3RFI
 
 | |
3QS1
 
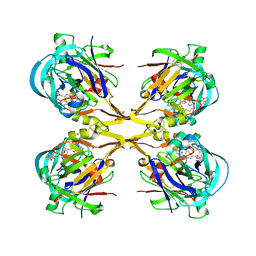 | | Crystal structure of KNI-10006 complex of Plasmepsin I (PMI) from Plasmodium falciparum | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-dimethylphenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Plasmepsin-1 | | Authors: | Bhaumik, P, Gustchina, A, Wlodawer, A. | | Deposit date: | 2011-02-19 | | Release date: | 2011-05-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the free and inhibited forms of plasmepsin I (PMI) from Plasmodium falciparum.
J.Struct.Biol., 175, 2011
|
|
2GD0
 
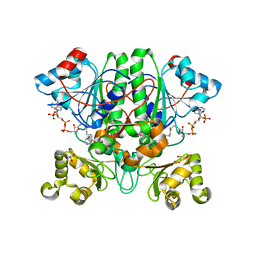 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (S)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD6
 
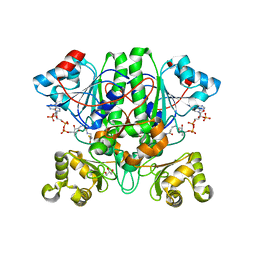 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GCE
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-IBUPROFENOYL-COENZYME A, (S)-IBUPROFENOYL-COENZYME A, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GCI
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an asparte/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | (R)-2-METHYLMYRISTOYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-14 | | Release date: | 2007-02-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
2GD2
 
 | | The 1,1-proton transfer reaction mechanism by alpha-methylacyl-CoA racemase is catalyzed by an aspartate/histidine pair and involves a smooth, methionine-rich surface for binding the fatty acyl moiety | | Descriptor: | ACETOACETYL-COENZYME A, GLYCEROL, probable alpha-methylacyl-CoA racemase MCR | | Authors: | Bhaumik, P, Wierenga, R.K. | | Deposit date: | 2006-03-15 | | Release date: | 2007-02-20 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Catalysis of the 1,1-Proton Transfer by alpha-Methyl-acyl-CoA Racemase Is Coupled to a Movement of the Fatty Acyl Moiety Over a Hydrophobic, Methionine-rich Surface
J.Mol.Biol., 367, 2007
|
|
9IQB
 
 | |
8GOG
 
 | | Structure of streptavidin mutant (S112Y-K121E) complexed with biotin-cyclopentadienyl-rhodium (III)(Cp*-Rh(III)) | | Descriptor: | CHLORIDE ION, GLYCEROL, RHODIUM(III) ION, ... | | Authors: | Sairaman, A, Mukherjee, P, Maiti, D, Bhaumik, P. | | Deposit date: | 2022-08-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiodivergent synthesis of isoindolones catalysed by a Rh(III)-based artificial metalloenzyme
Nat Synth, 3, 2024
|
|
2YIM
 
 | | The enolisation chemistry of a thioester-dependent racemase: the 1.4 A crystal structure of a complex with a planar reaction intermediate analogue | | Descriptor: | 2-METHYLACETOACETYL COA, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Sharma, S, Bhaumik, P, Venkatesan, R, Hiltunen, J.K, Conzelmann, E, Juffer, A.H, Wierenga, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | The Enolization Chemistry of a Thioester-Dependent Racemase: The 1.4 A Crystal Structure of a Reaction Intermediate Complex Characterized by Detailed Qm/Mm Calculations.
J Phys Chem B, 116, 2012
|
|
5XVX
 
 | | Crystal Structure of Aspergillus niger Glutamate Dehydrogenase Complexed With Alpha-ketoglutarate and NADPH | | Descriptor: | 2-OXOGLUTARIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2017-06-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the catalytic mechanism and alpha-ketoglutarate cooperativity of glutamate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
5XVI
 
 | |
5XWC
 
 | | Crystal Structure of Aspergillus niger Glutamate Dehydrogenase Complexed With Alpha-iminoglutarate, 2-amino-2-hydroxyglutarate and NADP | | Descriptor: | (2S)-2-azanyl-2-oxidanyl-pentanedioic acid, (2Z)-2-iminopentanedioic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2017-06-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the catalytic mechanism and alpha-ketoglutarate cooperativity of glutamate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
5XW0
 
 | | Crystal Structure of Aspergillus niger Glutamate Dehydrogenase Complexed With Isophthalate and NADPH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Glutamate dehydrogenase, ... | | Authors: | Prakash, P, Punekar, N.S, Bhaumik, P. | | Deposit date: | 2017-06-28 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the catalytic mechanism and alpha-ketoglutarate cooperativity of glutamate dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
9J4Z
 
 | |
9J50
 
 | |
9J4Y
 
 | |
