6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
4X46
 
 | | X-RAY structure thymidine phosphorylase from Salmonella typhimurium complex with SO4 at 2.19 A | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Thymidine phosphorylase | | Authors: | Balaev, V.V, Lashkov, A.A, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-RAY structure thymidine phosphorylase from Salmonella typhimurium complex with SO4 at 2.19 A
To Be Published
|
|
4YYY
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with uridine | | Descriptor: | CITRIC ACID, TRIETHYLENE GLYCOL, Thymidine phosphorylase, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-03-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural investigation of the thymidine phosphorylase from Salmonella typhimurium in the unliganded state and its complexes with thymidine and uridine.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5EPU
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytosine at 1.06A.
To Be Published
|
|
5EY3
 
 | | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with cytidine and sulphate | | Descriptor: | 1,2-ETHANEDIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, GLYCEROL, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Gabdulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-24 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | X-ray structure of the thymidine phosphorylase from Salmonella typhimurium in complex with cytidine and sulphate
To Be Published
|
|
6FDG
 
 | |
5EFO
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with cytidine and cytosine at 1.63A. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-10-24 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uridine at 2.24 A resolution
To Be Published
|
|
1FV0
 
 | | FIRST STRUCTURAL EVIDENCE OF THE INHIBITION OF PHOSPHOLIPASE A2 BY ARISTOLOCHIC ACID: CRYSTAL STRUCTURE OF A COMPLEX FORMED BETWEEN PHOSPHOLIPASE A2 AND ARISTOLOCHIC ACID | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 9-HYDROXY ARISTOLOCHIC ACID, ACETATE ION, ... | | Authors: | Chandra, V, Jasti, J, Kaur, P, Srinivasan, A, Betzel, C, Singh, T.P. | | Deposit date: | 2000-09-18 | | Release date: | 2002-08-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Phospholipase A2 Inhibition for the Synthesis of Prostaglandins by the Plant Alkaloid Aristolochic Acid from a 1.7 A Crystal Structure
Biochemistry, 41, 2002
|
|
5EP8
 
 | | X-Ray Structure of the Complex Pyrimidine-nucleoside phosphorylase from Bacillus subtilis with Sulfate Ion | | Descriptor: | Pyrimidine-nucleoside phosphorylase, SODIUM ION, SULFATE ION | | Authors: | Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2015-11-11 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | X-Ray Structure of the Complex Pyrimidine-nucleoside phosphorylase from Bacillus subtilis with Sulfate Ion
To Be Published
|
|
3R3L
 
 | | Structure of NP protein from Lassa AV strain | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, ZINC ION | | Authors: | Perbandt, M, Brunotte, L, Gunther, S, Betzel, C. | | Deposit date: | 2011-03-16 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Structure of the Lassa virus nucleoprotein revealed by X-ray crystallography, small-angle X-ray scattering, and electron microscopy.
J.Biol.Chem., 286, 2011
|
|
4FN6
 
 | | Structural Characterization of Thiaminase type II TenA from Staphylococcus aureus | | Descriptor: | ACETATE ION, GLYCEROL, thiaminase-2 | | Authors: | Begum, A, Drebes, J, Perbandt, M, Wrenger, C, Betzel, C. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural Characterization of Thiaminase type II TenA from Staphylococcus aureus
TO BE PUBLISHED
|
|
4N4Z
 
 | | Trypanosoma brucei procathepsin B structure solved by Serial Microcrystallography using synchrotron radiation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gati, C, Bourenkov, G, Klinge, M, Rehders, D, Stellato, F, Oberthuer, D, White, T.A, Yevanov, O, Sommer, B.P, Mogk, S, Duszenko, M, Betzel, C, Schneider, T.R, Chapman, H.N, Redecke, L. | | Deposit date: | 2013-10-08 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Serial crystallography on in vivo grown microcrystals using synchrotron radiation.
IUCrJ, 1, 2014
|
|
4O34
 
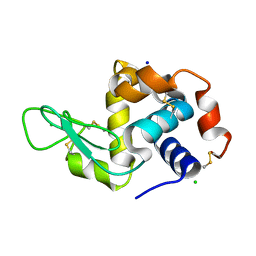 | | Room temperature macromolecular serial crystallography using synchrotron radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Stellato, F, Oberthuer, D, Liang, M, Bean, R, Gati, C, Yefanov, O, Barty, A, Burkhardt, A, Fischer, P, Galli, L, Kirian, R.A, Mayer, J, Pannerselvam, S, Yoon, C.H, Chervinskii, F, Speller, E, White, T.A, Betzel, C, Meents, A, Chapman, H.N. | | Deposit date: | 2013-12-18 | | Release date: | 2014-06-11 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Room-temperature macromolecular serial crystallography using synchrotron radiation.
IUCrJ, 1, 2014
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
6Y0H
 
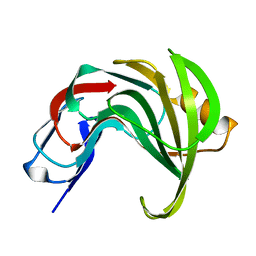 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
3G8F
 
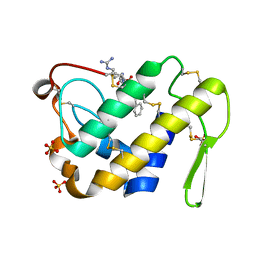 | | Crystal structure of the complex formed between a group II phospholipase A2 and designed peptide inhibitor carbobenzoxy-dehydro-val-ala-arg-ser at 1.2 A resolution | | Descriptor: | PHQ VAL ALA ARG SER peptide, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Kaur, P, Prem Kumar, R, Somvanshi, R.K, Perbandt, M, Betzel, C, Dey, S, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of the Complex Formed between a Group II Phospholipase A2 and Designed Peptide Inhibitor Carbobenzoxy-Dehydro-Val-Ala-Arg-Ser at 1.2 A Resolution
To be Published
|
|
4OGL
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymine at 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-16 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with thymine at 1.25 A resolution
Crystallogr. Rep., 2016
|
|
6FJS
 
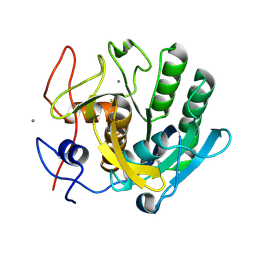 | | Proteinase~K SIRAS phased structure of room-temperature, serially collected synchrotron data | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Botha, S, Baitan, D, Jungnickel, K.E.J, Oberthuer, D, Schmidt, C, Stern, S, Wiedorn, M.O, Perbandt, M, Chapman, H.N, Betzel, C. | | Deposit date: | 2018-01-23 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novoprotein structure determination by heavy-atom soaking in lipidic cubic phase and SIRAS phasing using serial synchrotron crystallography.
IUCrJ, 5, 2018
|
|
2XSL
 
 | | The crystal structure of a Thermus thermophilus tRNAGly acceptor stem microhelix at 1.6 Angstroem resolution | | Descriptor: | 5'-R(*CP*UP*CP*CP*CP*GP*C)-3', 5'-R(*GP*CP*GP*GP*GP*AP*G)-3' | | Authors: | Oberthuer, D, Eichert, A, Erdmann, V.A, Fuerste, J.P, Betzel, C, Foerster, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The Crystal Structure of a Thermus Thermophilus tRNA(Gly) Acceptor Stem Microhelix at 1.6 A Resolution.
Biochem.Biophys.Res.Commun., 404, 2011
|
|
2X2Q
 
 | | Crystal structure of an 'all locked' LNA duplex at 1.9 angstrom resolution | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), LOCKED NUCLEIC ACID DERIVED FROM TRNA SER ACCEPTOR STEM MICROHELIX, ... | | Authors: | Eichert, A, Behling, K, Fuerste, J.P, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2010-01-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of an 'All Locked' Nucleic Acid Duplex.
Nucleic Acids Res., 38, 2010
|
|
2W89
 
 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
4OEH
 
 | | X-ray Structure of Uridine Phosphorylase from Vibrio cholerae Complexed with Uracil at 1.91 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ETHANOL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | X-ray structures of uridine phosphorylase from Vibrio cholerae in complexes with uridine, thymidine, uracil, thymine, and phosphate anion: Substrate specificity of bacterial uridine phosphorylases.
To Be Published, Crystallogr. Rep., 2016
|
|
2V6W
 
 | | tRNASer acceptor stem: Conformation and hydration of a microhelix in a crystal structure at 1.8 Angstrom resolution | | Descriptor: | 5'-R(*GP*GP*AP*GP*AP*GP*AP)-3', 5'-R(*UP*CP*UP*CP*UP*CP*CP)-3' | | Authors: | Foerster, C, Brauer, A.B.E, Brode, S, Fuerste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-07-23 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Trnaser Acceptor Stem: Conformation and Hydration of a Microhelix in a Crystal Structure at 1.8 A Resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3H9D
 
 | | Crystal Structure of Trypanosoma brucei ATG8 | | Descriptor: | CALCIUM ION, Microtubule-associated protein 1A/1B, light chain 3, ... | | Authors: | Koopmann, R, Muhammad, K, Perbandt, M, Betzel, C, Duszenko, M. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Trypanosoma brucei ATG8: structural insights into autophagic-like mechanisms in protozoa.
Autophagy, 5, 2009
|
|
4QXD
 
 | | Crystal structure of Inositol Polyphosphate 1-Phosphatase from Entamoeba histolytica | | Descriptor: | 3'(2'),5'-bisphosphate nucleotidase, putative, MAGNESIUM ION, ... | | Authors: | Tarique, K.F, Abdul Rehman, S.A, Betzel, C, Gourinath, S. | | Deposit date: | 2014-07-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-based identification of inositol polyphosphate 1-phosphatase from Entamoeba histolytica
Acta Crystallogr.,Sect.D, 70, 2014
|
|
