388D
 
 | | CRYSTAL STRUCTURE OF B-DNA WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINES: IMPLICATIONS OF CONFORMATIONAL PREORGANIZATION FOR DUPLEX STABILITY | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*(TAF)P*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Berger, I, Tereshko, V, Ikeda, H, Marquez, V.E, Egli, M. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of B-DNA with incorporated 2'-deoxy-2'-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability.
Nucleic Acids Res., 26, 1998
|
|
389D
 
 | | CRYSTAL STRUCTURE OF B-DNA WITH INCORPORATED 2'-DEOXY-2'-FLUORO-ARABINO-FURANOSYL THYMINES: IMPLICATIONS OF CONFORMATIONAL PREORGANIZATION FOR DUPLEX STABILITY | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(TAF)P*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Berger, I, Tereshko, V, Ikeda, H, Marquez, V.E, Egli, M. | | Deposit date: | 1998-04-20 | | Release date: | 1998-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structures of B-DNA with incorporated 2'-deoxy-2'-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability.
Nucleic Acids Res., 26, 1998
|
|
1IMS
 
 | | MOLECULAR STRUCTURE OF THE HALOGENATED ANTI-CANCER DRUG IODODOXORUBICIN COMPLEXED WITH D(TGTACA) AND D(CGATCG) | | Descriptor: | 4'-DEOXY-4'-IODODOXORUBICIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Berger, I, Su, L, Spitzner, J.R, Kang, C, Burke, T.G, Rich, A. | | Deposit date: | 1995-10-23 | | Release date: | 1996-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular structure of the halogenated anti-cancer drug iododoxorubicin complexed with d(TGTACA) and d(CGATCG).
Nucleic Acids Res., 23, 1995
|
|
1IMR
 
 | | MOLECULAR STRUCTURE OF THE HALOGENATED ANTI-CANCER DRUG IODODOXORUBICIN COMPLEXED WITH D(TGTACA) AND D(CGATCG) | | Descriptor: | 4'-DEOXY-4'-IODODOXORUBICIN, DNA (5'-D(*TP*GP*TP*AP*CP*A)-3') | | Authors: | Berger, I, Su, L, Spitzner, J.R, Kang, C, Burke, T.G, Rich, A. | | Deposit date: | 1995-10-23 | | Release date: | 1996-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular structure of the halogenated anti-cancer drug iododoxorubicin complexed with d(TGTACA) and d(CGATCG).
Nucleic Acids Res., 23, 1995
|
|
241D
 
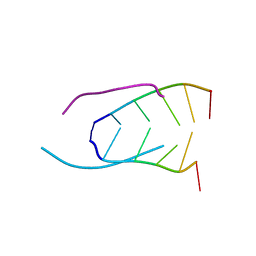 | | EXTENSION OF THE FOUR-STRANDED INTERCALATED CYTOSINE MOTIF BY ADENINE.ADENINE BASE PAIRING IN THE CRYSTAL STRUCTURE OF D(CCCAAT) | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*AP*T)-3') | | Authors: | Berger, I, Kang, C, Fredian, A, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extension of the four-stranded intercalated cytosine motif by adenine.adenine base pairing in the crystal structure of d(CCCAAT).
Nat.Struct.Biol., 2, 1995
|
|
8COI
 
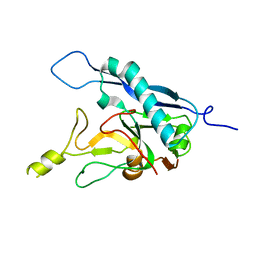 | | Human adenovirus-derived synthetic ADDobody binder | | Descriptor: | ADDobody | | Authors: | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8F2R
 
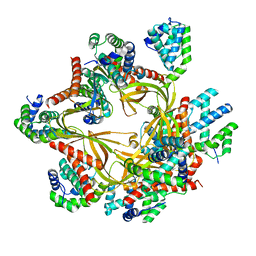 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8F2U
 
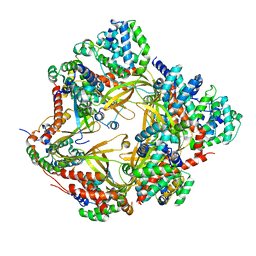 | | Human CCC complex | | Descriptor: | COMM domain-containing protein 1, COMM domain-containing protein 10, COMM domain-containing protein 2, ... | | Authors: | Healy, M.D, McNally, K.E, Butkovic, R, Chilton, M, Kato, K, Sacharz, J, McConville, C, Moody, E.R.R, Shaw, S, Planelles-Herrero, V.J, Kadapalakere, S.Y, Ross, J, Borucu, U, Palmer, C.S, Chen, K, Croll, T.I, Hall, R.J, Caruana, N.J, Ghai, R, Nguyen, T.H.D, Heesom, K.J, Saitoh, S, Berger, I, Berger-Schaffitzel, C, Williams, T.A, Stroud, D.A, Derivery, E, Collins, B.M, Cullen, P.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
6F3T
 
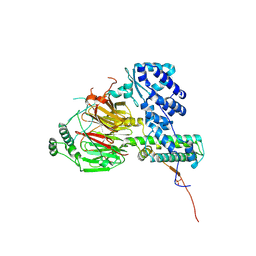 | | Crystal structure of the human TAF5-TAF6-TAF9 complex | | Descriptor: | CHLORIDE ION, Transcription initiation factor TFIID subunit 5, Transcription initiation factor TFIID subunit 6, ... | | Authors: | Haffke, M, Berger, I. | | Deposit date: | 2017-11-28 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chaperonin CCT checkpoint function in basal transcription factor TFIID assembly.
Nat. Struct. Mol. Biol., 25, 2018
|
|
8C9N
 
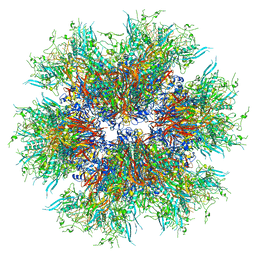 | | MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle | | Descriptor: | Penton protein | | Authors: | Bufton, J.C, Capin, J, Boruku, U, Garzoni, F, Schaffitzel, C, Berger, I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | In vitro generated antibodies guide thermostable ADDomer nanoparticle design for nasal vaccination and passive immunization against SARS-CoV-2.
Antib Ther, 6, 2023
|
|
7ZH2
 
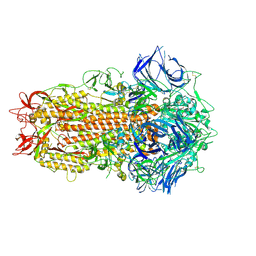 | | SARS CoV Spike protein, Closed C1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH5
 
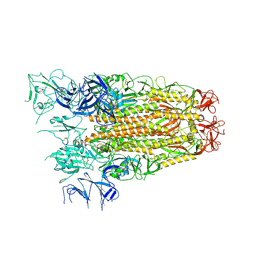 | | SARS CoV Spike protein, Open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH1
 
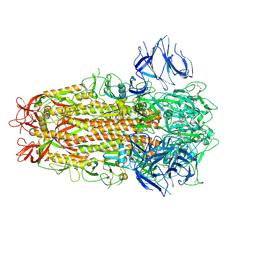 | | SARS CoV Spike protein, Closed C3 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
9H1M
 
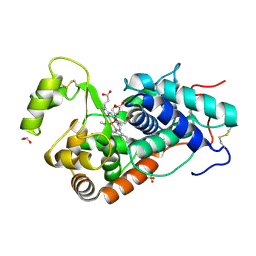 | | Recombinant ferric horseradish peroxidase C1A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Nesa, M.L, Mandal, S.K, Toelzer, C, Humer, D, Moody, P.C.E, Berger, I, Spadiut, O, Raven, E.L. | | Deposit date: | 2024-10-09 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of ferric recombinant horseradish peroxidase.
J.Biol.Inorg.Chem., 30, 2025
|
|
3ZN8
 
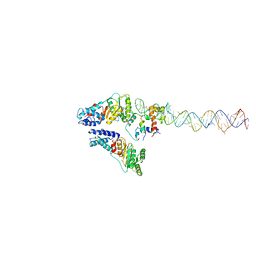 | | Structural Basis of Signal Sequence Surveillance and Selection by the SRP-SR Complex | | Descriptor: | 4.5 S RNA, DIPEPTIDYL AMINOPEPTIDASE B, MAGNESIUM ION, ... | | Authors: | von Loeffelholz, O, Knoops, K, Ariosa, A, Zhang, X, Karuppasamy, M, Huard, K, Schoehn, G, Berger, I, Shan, S.O, Schaffitzel, C. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural Basis of Signal Sequence Surveillance and Selection by the Srp-Sr Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
8QB3
 
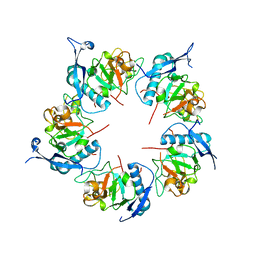 | | ADDobody zinc containing condition | | Descriptor: | ADDobody, ZINC ION | | Authors: | Buzas, D, Toelzer, C, Berger, I, Schaffitzel, C. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
6HCR
 
 | | Synthetic Self-assembling ADDomer Platform for Highly Efficient Vaccination by Genetically-encoded Multi-epitope Display | | Descriptor: | Penton protein | | Authors: | Bufton, J.C, Berger, I, Schaffitzel, C, Garzoni, F, Rabi, F.A. | | Deposit date: | 2018-08-16 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Synthetic self-assembling ADDomer platform for highly efficient vaccination by genetically encoded multiepitope display.
Sci Adv, 5, 2019
|
|
7OD3
 
 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7ODL
 
 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
2IY3
 
 | | Structure of the E. Coli Signal Regognition Particle | | Descriptor: | 4.5S RNA, SIGNAL SEQUENCE, Signal recognition particle protein,Signal recognition particle 54 kDa protein | | Authors: | Schaffitzel, C, Oswald, M, Berger, I, Ishikawa, T, Abrahams, J.P, Koerten, H.K, Koning, R.I, Ban, N. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structure of the E. Coli Signal Recognition Particle Bound to a Translating Ribosome
Nature, 444, 2006
|
|
3RJ1
 
 | | Architecture of the Mediator Head module | | Descriptor: | Mediator of RNA polymerase II transcription subunit 11, Mediator of RNA polymerase II transcription subunit 17, Mediator of RNA polymerase II transcription subunit 18, ... | | Authors: | Imasaki, T, Calero, G, Cai, G, Tsai, K.L, Yamada, K, Cardelli, F, Erdjument-Bromage, H, Tempst, P, Berger, I, Kornberg, G.L, Asturias, F.J, Kornberg, R.D, Takagi, Y. | | Deposit date: | 2011-04-14 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Architecture of the Mediator head module.
Nature, 475, 2011
|
|
3U3K
 
 | | Crystal structure of hSULT1A1 bound to PAP and 2-Naphtol | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1, naphthalen-2-ol | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3U3R
 
 | | Crystal structure of D249G mutated Human SULT1A1 bound to PAP and P-NITROPHENOL | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, P-NITROPHENOL, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3U3J
 
 | | Crystal structure of hSULT1A1 bound to PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
3U3M
 
 | | Crystal structure of Human SULT1A1 bound to PAP and 3-Cyano-7-hydroxycoumarin | | Descriptor: | 7-hydroxy-2-oxo-2H-chromene-3-carbonitrile, ADENOSINE-3'-5'-DIPHOSPHATE, Sulfotransferase 1A1 | | Authors: | Guttman, C, Berger, I, Aharoni, A, Zarivach, R. | | Deposit date: | 2011-10-06 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The molecular basis for the broad substrate specificity of human sulfotransferase 1A1.
Plos One, 6, 2011
|
|
