2ND6
 
 | | Structure of DK17 in GM1 LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND7
 
 | | Structure of DK17 in POPC:POPG:Cholesterol:GM1 LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND8
 
 | | Structures of DK17 in TBLE LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
7YYK
 
 | | Crystal structure of the O-fucosylated form of TSRs1-3 from the human thrombospondin 1 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1, alpha-L-fucopyranose | | Authors: | Berardinelli, S.J, Eletsky, A, Valero-Gonzalez, J, Ito, A, Manjunath, R, Hurtado-Guerrero, R, Prestegard, J.R, Woods, R.J, Haltiwanger, R.S. | | Deposit date: | 2022-02-18 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | O-fucosylation stabilizes the TSR3 motif in thrombospondin-1 by interacting with nearby amino acids and protecting a disulfide bond.
J.Biol.Chem., 298, 2022
|
|
1U5K
 
 | | Recombinational repair protein RecO | | Descriptor: | ZINC ION, hypothetical protein | | Authors: | Makharashvili, N, Koroleva, O, Bera, S, Grandgenett, D.P, Korolev, S. | | Deposit date: | 2004-07-27 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel Structure of DNA Repair Protein RecO from Deinococcus radiodurans
STRUCTURE, 12, 2004
|
|
7JN3
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048 | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KU7
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. Cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KUI
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. CIC region of a cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
8E14
 
 | | Cryo-EM structure of Rous sarcoma virus strand transfer complex | | Descriptor: | DNA (42-MER), DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*TP*AP*CP*TP*C)-3'), DNA (5'-D(*AP*GP*TP*GP*TP*CP*TP*TP*CP*TP*TP*CP*TP*TP*TP*C)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2022-08-09 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Molecular determinants for Rous sarcoma virus intasome assemblies involved in retroviral integration.
J.Biol.Chem., 299, 2023
|
|
8A59
 
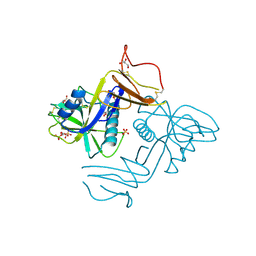 | | C-type lectin-like domain (CTLD) and Sushi-like domain of human CD93 | | Descriptor: | Complement component C1q receptor, GLYCEROL, SULFATE ION | | Authors: | Tassone, G, Barbera, S, Raucci, L, Orlandini, M, Pozzi, C. | | Deposit date: | 2022-06-14 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Dimerization of the C-type lectin-like receptor CD93 promotes its binding to Multimerin-2 in endothelial cells.
Int.J.Biol.Macromol., 224, 2023
|
|
4BIH
 
 | | Crystal structure of the conserved staphylococcal antigen 1A, Csa1A | | Descriptor: | CALCIUM ION, UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Spraggon, G, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.459 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4BIG
 
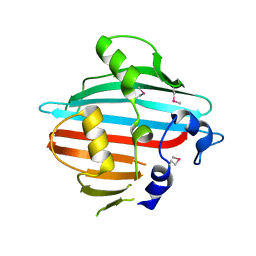 | | Crystal structure of the conserved staphylococcal antigen 1B, Csa1B | | Descriptor: | UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | Authors: | Malito, E, Bottomley, M.J, Schluepen, C, Liberatori, S. | | Deposit date: | 2013-04-10 | | Release date: | 2013-08-07 | | Last modified: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
