1QZV
 
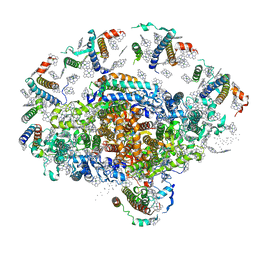 | | Crystal structure of plant photosystem I | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHYLLOQUINONE, ... | | Authors: | Ben-Shem, A, Frolow, F, Nelson, N. | | Deposit date: | 2003-09-18 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.44 Å) | | Cite: | Crystal structure of plant photosystem I.
Nature, 426, 2003
|
|
4V7R
 
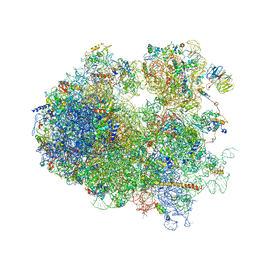 | | Yeast 80S ribosome. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Ben-Shem, A, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2010-07-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of the eukaryotic ribosome.
Science, 330, 2010
|
|
4V88
 
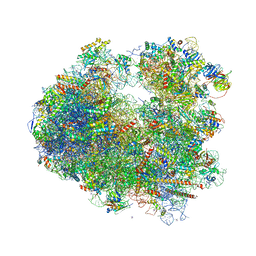 | | The structure of the eukaryotic ribosome at 3.0 A resolution. | | Descriptor: | 18S RIBOSOMAL RNA, 18S rRNA, 25S rRNA, ... | | Authors: | Ben-Shem, A, Garreau de Loubresse, N, Melnikov, S, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2011-10-11 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the eukaryotic ribosome at 3.0 angstrom resolution.
Science, 334, 2011
|
|
7ZVW
 
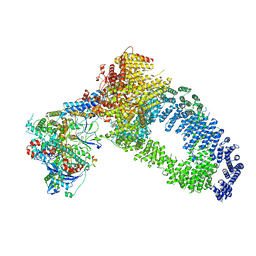 | | NuA4 Histone Acetyltransferase Complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Chromatin modification-related protein EAF1, ... | | Authors: | Schultz, P, Ben-Shem, A, Frechard, A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-31 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structure of the NuA4-Tip60 complex reveals the mechanism and importance of long-range chromatin modification.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OF4
 
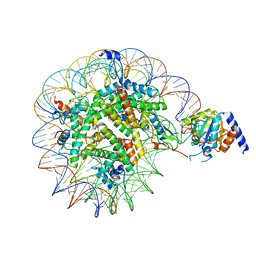 | | Nucleosome Bound human SIRT6 (Composite) | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Smirnova, E, Bignon, E, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Binding to nucleosome poises human SIRT6 for histone H3 deacetylation.
Elife, 12, 2024
|
|
6HQA
 
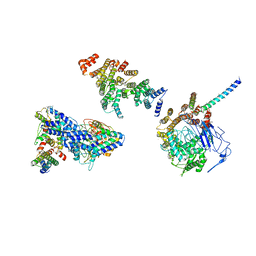 | | Molecular structure of promoter-bound yeast TFIID | | Descriptor: | Histone-fold, Subunit (17 kDa) of TFIID and SAGA complexes, involved in RNA polymerase II transcription initiation, ... | | Authors: | Kolesnikova, O, Ben-Shem, A, Luo, J, Ranish, J, Schultz, P, Papai, G. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular structure of promoter-bound yeast TFIID.
Nat Commun, 9, 2018
|
|
8QR1
 
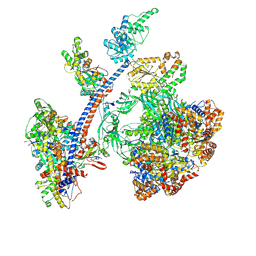 | | Cryo-EM structure of the human Tip60 complex | | Descriptor: | Actin, cytoplasmic 1, N-terminally processed, ... | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 2024
|
|
8QRI
 
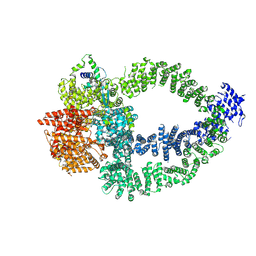 | | TRRAP and EP400 in the human Tip60 complex | | Descriptor: | E1A-binding protein p400, Transformation/transcription domain-associated protein | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, SChultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-09 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the human TIP60-C histone exchange and acetyltransferase complex.
Nature, 2024
|
|
2IRV
 
 | | Crystal structure of GlpG, a rhomboid intramembrane serine protease | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, DODECYL-BETA-D-MALTOSIDE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Bibi, E, Fass, D, Ben-Shem, A. | | Deposit date: | 2006-10-16 | | Release date: | 2006-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for intramembrane proteolysis by rhomboid serine proteases.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6TB4
 
 | | Structure of SAGA bound to TBP | | Descriptor: | SAGA-associated factor 73 (Sgf73), Spt20, Subunit (17 kDa) of TFIID and SAGA complexes, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-10-31 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
6TBM
 
 | | Structure of SAGA bound to TBP, including Spt8 and DUB | | Descriptor: | Polyubiquitin-B, SAGA-associated factor 11, Spt20, ... | | Authors: | Papai, G, Frechard, A, Kolesnikova, O, Crucifix, C, Schultz, P, Ben-Shem, A. | | Deposit date: | 2019-11-01 | | Release date: | 2020-02-12 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Structure of SAGA and mechanism of TBP deposition on gene promoters.
Nature, 577, 2020
|
|
5OEJ
 
 | | Structure of Tra1 subunit within the chromatin modifying complex SAGA | | Descriptor: | Tra1 subunit within the chromatin modifying complex SAGA | | Authors: | Sharov, G, Voltz, K, Durand, A, Kolesnikova, O, Papai, G, Myasnikov, A.G, Dejaegere, A, Ben-Shem, A, Schultz, P. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure of the transcription activator target Tra1 within the chromatin modifying complex SAGA.
Nat Commun, 8, 2017
|
|
