7OOJ
 
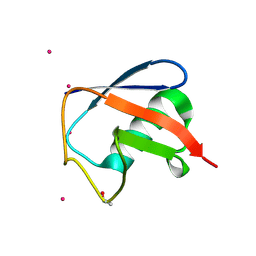 | | Structure of D-Thr53 Ubiquitin | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Becker, S. | | Deposit date: | 2021-05-27 | | Release date: | 2022-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A litmus test for classifying recognition mechanisms of transiently binding proteins.
Nat Commun, 13, 2022
|
|
7QV1
 
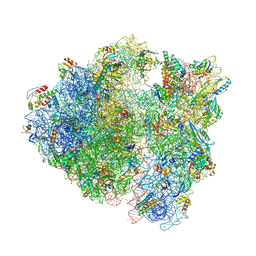 | | Bacillus subtilis collided disome (Leading 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
7QV2
 
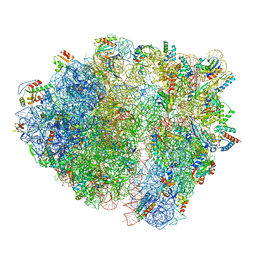 | | Bacillus subtilis collided disome (Collided 70S) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2022-01-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Bacterial ribosome collision sensing by a MutS DNA repair ATPase paralogue.
Nature, 603, 2022
|
|
8BJP
 
 | |
1BG1
 
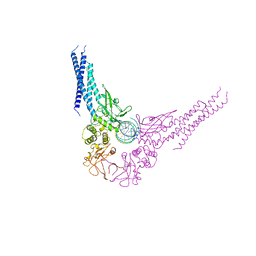 | | TRANSCRIPTION FACTOR STAT3B/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*GP*CP*AP*TP*TP*TP*CP*CP*CP*GP*TP*AP*AP*AP*TP*CP*T)-3'), PROTEIN (TRANSCRIPTION FACTOR STAT3B) | | Authors: | Becker, S, Groner, B, Muller, C.W. | | Deposit date: | 1998-06-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Three-dimensional structure of the Stat3beta homodimer bound to DNA.
Nature, 394, 1998
|
|
5C58
 
 | |
8BGB
 
 | |
8BIY
 
 | |
7QV3
 
 | |
7AQD
 
 | |
7AQC
 
 | | Structure of the bacterial RQC complex (Decoding State) | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Filbeck, S, Pfeffer, S. | | Deposit date: | 2020-10-21 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Mimicry of Canonical Translation Elongation Underlies Alanine Tail Synthesis in RQC.
Mol.Cell, 81, 2021
|
|
2YBO
 
 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-08 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
2YBQ
 
 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
1RXR
 
 | | HIGH RESOLUTION SOLUTION STRUCTURE OF THE RETINOID X RECEPTOR DNA BINDING DOMAIN, NMR, 20 STRUCTURE | | Descriptor: | RETINOIC ACID RECEPTOR-ALPHA, ZINC ION | | Authors: | Holmbeck, S.M.A, Foster, M.P, Casimiro, D.R, Sem, D.S, Dyson, H.J, Wright, P.E. | | Deposit date: | 1998-06-12 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of the retinoid X receptor DNA-binding domain.
J.Mol.Biol., 281, 1998
|
|
6FCG
 
 | |
5M28
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to maltotriose | | Descriptor: | CHLORIDE ION, MalE1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2016-10-12 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5MK9
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to beta-cyclodextrin | | Descriptor: | CHLORIDE ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2016-12-02 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5MKB
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 without ligand | | Descriptor: | MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2016-12-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5MKA
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to gamma-cyclodextrin | | Descriptor: | Cyclooctakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2016-12-02 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.149 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5MTU
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to alpha-cyclodextrin | | Descriptor: | Cyclohexakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.999 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
5MTT
 
 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to maltotetraose | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MalE1, ... | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2017-01-10 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
4MHX
 
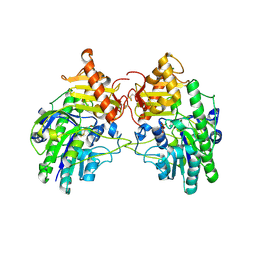 | | Crystal Structure of Sulfamidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sidhu, N.S, Uson, I, Schreiber, K, Proepper, K, Becker, S, Gaertner, J, Kraetzner, R, Steinfeld, R, Sheldrick, G.M. | | Deposit date: | 2013-08-30 | | Release date: | 2014-05-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of sulfamidase provides insight into the molecular pathology of mucopolysaccharidosis IIIA.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1G8X
 
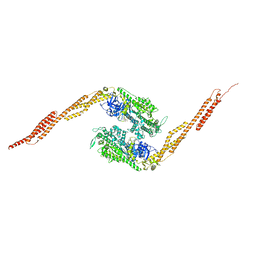 | | STRUCTURE OF A GENETICALLY ENGINEERED MOLECULAR MOTOR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN FUSED TO ALPHA-ACTININ 3 | | Authors: | Kliche, W, Fujita-Becker, S, Kollmar, M, Manstein, D.J, Kull, F.J. | | Deposit date: | 2000-11-21 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a genetically engineered molecular motor.
EMBO J., 20, 2001
|
|
1LO1
 
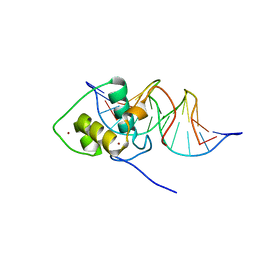 | | ESTROGEN RELATED RECEPTOR 2 DNA BINDING DOMAIN IN COMPLEX WITH DNA | | Descriptor: | 5'-D(*CP*GP*TP*GP*AP*CP*CP*TP*TP*GP*AP*GP*C)-3', 5'-D(*GP*CP*TP*CP*AP*AP*GP*GP*TP*CP*AP*CP*G)-3', Steroid hormone receptor ERR2, ... | | Authors: | Gearhart, M.D, Holmbeck, S.M.A, Evans, R.M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-05-05 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Monomeric Complex of Human Orphan Estrogen Related Receptor-2 with DNA: A Pseudo-dimer Interface Mediates Extended Half-site Recognition
J.Mol.Biol., 327, 2003
|
|
8A4L
 
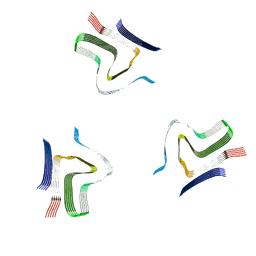 | | Lipidic alpha-synuclein fibril - polymorph L2A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-06-12 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
