6ENS
 
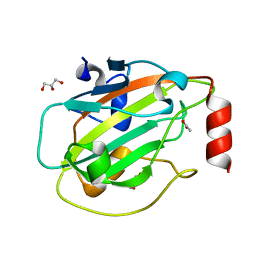 | | Structure of mouse wild-type RKIP | | Descriptor: | ACETATE ION, GLYCEROL, Phosphatidylethanolamine-binding protein 1 | | Authors: | Hirschbeck, M, Koelmel, W, Schindelin, H, Lorenz, K, Kisker, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Conserved salt-bridge competition triggered by phosphorylation regulates the protein interactome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2N64
 
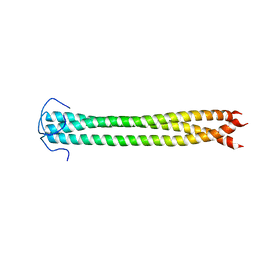 | |
4DC7
 
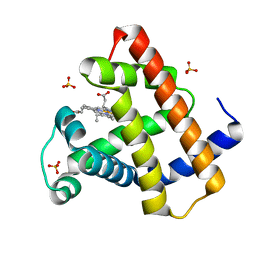 | | Crystal Structure of Myoglobin Exposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Becker, M, Mulichak, A.M, Kissick, D.J, Fischetti, R.F, Keefe, L.J, Simpson, G.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2VFO
 
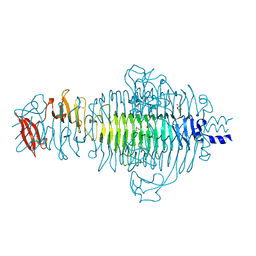 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V125L | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFQ
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V450A | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN,, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFN
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V125A | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFP
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666), Mutant V349L | | Descriptor: | CALCIUM ION, GLYCEROL, P22 TAILSPIKE PROTEIN, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
2VFM
 
 | | Low Temperature Structure of P22 Tailspike Protein Fragment (109-666) | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, CALCIUM ION, GLYCEROL, ... | | Authors: | Becker, M, Mueller, J.J, Heinemann, U, Seckler, R. | | Deposit date: | 2007-11-05 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Side-Chain Stacking and Beta-Helix Stability in P22 Tailspike Protein
To be Published
|
|
1P4P
 
 | | Outer Surface Protein B of B. burgdorferi: crystal structure of the C-terminal fragment | | Descriptor: | Outer surface protein B | | Authors: | Becker, M, Bunikis, J, Lade, B.D, Dunn, J.J, Barbour, A.G, Lawson, C.L. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Investigation of Borrelia burgdorferi OspB, a BactericidalFab Target.
J.Biol.Chem., 280, 2005
|
|
1RJL
 
 | | Structure of the complex between OspB-CT and bactericidal Fab-H6831 | | Descriptor: | Fab H6831 H-chain, Fab H6831 L-chain, Outer surface protein B | | Authors: | Becker, M, Bunikis, J, Lade, B.D, Dunn, J.J, Barbour, A.G, Lawson, C.L. | | Deposit date: | 2003-11-19 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Investigation of Borrelia burgdorferi OspB, a BactericidalFab Target.
J.Biol.Chem., 280, 2005
|
|
4DC8
 
 | | Crystal Structure of Myoglobin Unexposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Becker, M, Mulichak, A.M, Kissick, D.J, Fischetti, R.F, Keefe, L.J, Simpson, D.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2LGC
 
 | |
7OZS
 
 | | Structure of the hexameric 5S RNP from C. thermophilum | | Descriptor: | 5S rRNA, 60S ribosomal protein l5-like protein, Putative ribosomal protein, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2021-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 2023
|
|
7R5K
 
 | | Human nuclear pore complex (constricted) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores.
Science, 376, 2022
|
|
7R5J
 
 | | Human nuclear pore complex (dilated) | | Descriptor: | Aladin, E3 SUMO-protein ligase RanBP2, Nuclear pore complex protein Nup107, ... | | Authors: | Mosalaganti, S, Obarska-Kosinska, A, Siggel, M, Taniguchi, R, Turonova, B, Zimmerli, C.E, Buczak, K, Schmidt, F.H, Margiotta, E, Mackmull, M.T, Hagen, W.J.H, Hummer, G, Kosinski, J, Beck, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-09-21 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (50 Å) | | Cite: | AI-based structure prediction empowers integrative structural analysis of human nuclear pores
Science, 376, 2022
|
|
5A1U
 
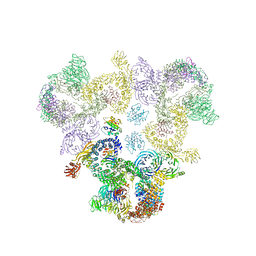 | | The structure of the COPI coat triad | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1Y
 
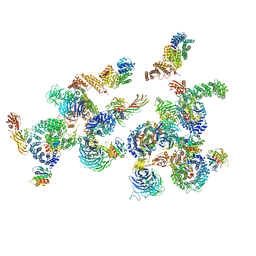 | | The structure of the COPI coat linkage IV | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1X
 
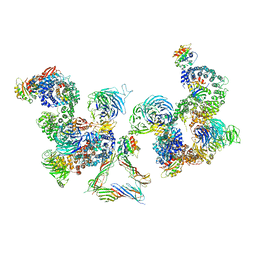 | | The structure of the COPI coat linkage III | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1V
 
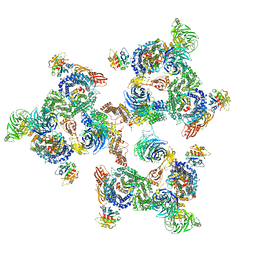 | | The structure of the COPI coat linkage I | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
5A1W
 
 | | The structure of the COPI coat linkage II | | Descriptor: | ADP-RIBOSYLATION FACTOR 1, COATOMER SUBUNIT ALPHA, COATOMER SUBUNIT BETA, ... | | Authors: | Dodonova, S.O, Diestelkoetter-Bachert, P, von Appen, A, Hagen, W.J.H, Beck, R, Beck, M, Wieland, F, Briggs, J.A.G. | | Deposit date: | 2015-05-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Vesicular Transport. A Structure of the Copi Coat and the Role of Coat Proteins in Membrane Vesicle Assembly.
Science, 349, 2015
|
|
7R1Y
 
 | |
8BGU
 
 | | human MDM2-5S RNP | | Descriptor: | 5S rRNA, 60S ribosomal protein L11, 60S ribosomal protein L5, ... | | Authors: | Castillo, N, Thoms, M, Flemming, D, Hammaren, H.M, Buschauer, R, Ameismeier, M, Bassler, J, Beck, M, Beckmann, R, Hurt, E. | | Deposit date: | 2022-10-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of nascent 5S RNPs at the crossroad between ribosome assembly and MDM2-p53 pathways.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5IJO
 
 | | Alternative composite structure of the inner ring of the human nuclear pore complex (16 copies of Nup188, 16 copies of Nup205) | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Kosinski, J, Mosalaganti, S, von Appen, A, Beck, M. | | Deposit date: | 2016-03-02 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21.4 Å) | | Cite: | Molecular architecture of the inner ring scaffold of the human nuclear pore complex.
Science, 352, 2016
|
|
5IJN
 
 | | Composite structure of the inner ring of the human nuclear pore complex (32 copies of Nup205) | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP155, NUCLEAR PORE COMPLEX PROTEIN NUP205, NUCLEAR PORE COMPLEX PROTEIN NUP54, ... | | Authors: | Kosinski, J, Mosalaganti, S, von Appen, A, Beck, M. | | Deposit date: | 2016-03-02 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (21.4 Å) | | Cite: | Molecular architecture of the inner ring scaffold of the human nuclear pore complex.
Science, 352, 2016
|
|
5A9Q
 
 | | Human nuclear pore complex | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | Authors: | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
