8R32
 
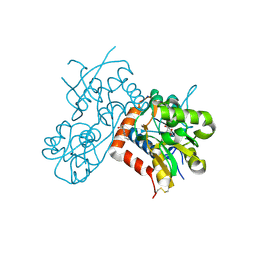 | | Crystal structure of the GluK2 ligand-binding domain in complex with L-glutamate and BPAM344 at 1.60 A resolution | | Descriptor: | 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, GLUTAMIC ACID, ... | | Authors: | Bay, Y, Jeppesen, M.E, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The positive allosteric modulator BPAM344 and L-glutamate introduce an active-like structure of the ligand-binding domain of GluK2.
Febs Lett., 598, 2024
|
|
8R36
 
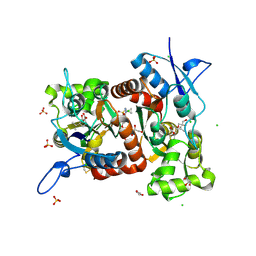 | | Crystal structure of the Gluk1 ligand-binding domain in complex with kainate and BPAM538 at 1.90 A resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Bay, Y, Frantsen, S.M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-11-08 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GluK1 ligand-binding domain with kainate and the full-spanning positive allosteric modulator BPAM538.
J.Struct.Biol., 216, 2024
|
|
6R9W
 
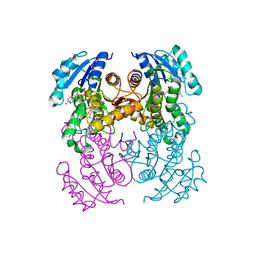 | | Crystal structure of InhA in complex with AP-124 inhibitor | | Descriptor: | (2~{S})-1-(benzimidazol-1-yl)-3-(2,3-dihydro-1~{H}-inden-5-yloxy)propan-2-ol, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takebayashi, Y, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-04-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of New and Potent InhA Inhibitors as Antituberculosis Agents: Structure-Based Virtual Screening Validated by Biological Assays and X-ray Crystallography.
J.Chem.Inf.Model., 60, 2020
|
|
6T1H
 
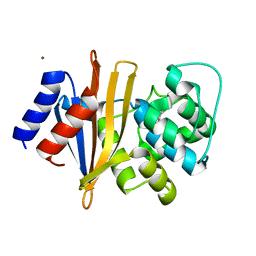 | | OXA-51-like beta-lactamase OXA-66 | | Descriptor: | Beta-lactamase OXA-66, ZINC ION | | Authors: | Takebayashi, Y, Chirgadze, D, Henderson, S, Warburton, P.J, Evans, B.E. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the OXA-51-like beta-lactamase OXA-66
To Be Published
|
|
7ZLL
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum in complex with the 5-[(morpholin-4-yl)methyl]quinolin-8-ol inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-(morpholin-4-ylmethyl)quinolin-8-ol, UDP-glucose-glycoprotein glucosyltransferase-like protein, ... | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Kantsadi, A.L, Chandran, A.V. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
1O5P
 
 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
1BQT
 
 | | THREE-DIMENSIONAL STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR-I (IGF-I) DETERMINED BY 1H-NMR AND DISTANCE GEOMETRY, 6 STRUCTURES | | Descriptor: | INSULIN-LIKE GROWTH FACTOR-I | | Authors: | Sato, A, Nishimura, S, Ohkubo, T, Kyogoku, Y, Koyama, S, Kobayashi, M, Yasuda, T, Kobayashi, Y. | | Deposit date: | 1998-08-18 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human insulin-like growth factor-I (IGF-I) determined by 1H-NMR and distance geometry.
Int.J.Pept.Protein Res., 41, 1993
|
|
7AAL
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1, G258A mutant | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAM
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 bound to the CTH domain of the phosphatase LYP | | Descriptor: | GLYCEROL, Proline-serine-threonine phosphatase-interacting protein 1, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
7AAN
 
 | | Crystal structure of the F-BAR domain of PSTIPIP1 | | Descriptor: | Proline-serine-threonine phosphatase-interacting protein 1 | | Authors: | Manso, J.A, Alcon, P, Bayon, Y, Alonso, A, de Pereda, J.M. | | Deposit date: | 2020-09-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | PSTPIP1-LYP phosphatase interaction: structural basis and implications for autoinflammatory disorders.
Cell.Mol.Life Sci., 79, 2022
|
|
5YQ0
 
 | | Crystal structure of secreted protein CofJ from ETEC. | | Descriptor: | CALCIUM ION, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YPZ
 
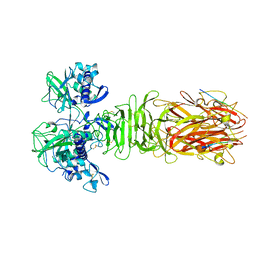 | | Crystal structure of minor pilin CofB from CFA/III complexed with N-terminal peptide fragment of CofJ | | Descriptor: | CofB, CofJ | | Authors: | Oki, H, Kawahara, K, Maruno, T, Imai, T, Muroga, Y, Fukakusa, S, Iwashita, T, Kobayashi, Y, Matsuda, S, Kodama, T, Iida, T, Yoshida, T, Ohkubo, T, Nakamura, S. | | Deposit date: | 2017-11-04 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.521 Å) | | Cite: | Interplay of a secreted protein with type IVb pilus for efficient enterotoxigenicEscherichia colicolonization.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4U4P
 
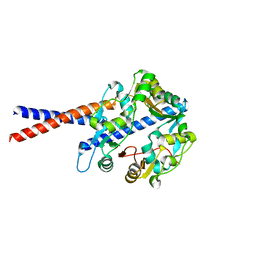 | | Crystal structure of the human condensin SMC hinge domain heterodimer with short coiled coils | | Descriptor: | Structural maintenance of chromosomes protein 2, Structural maintenance of chromosomes protein 4 | | Authors: | Uchiyama, S, Kawahara, K, Hosokawa, Y, Fukakusa, S, Oki, H, Nakamura, S, Noda, M, Takino, R, Miyahara, Y, Maruno, T, Kobayashi, Y, Ohkubo, T, Fukui, K. | | Deposit date: | 2014-07-24 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for dimer information and DNA recognition of human SMC proteins
to be published
|
|
8JLB
 
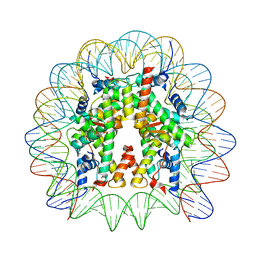 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JL9
 
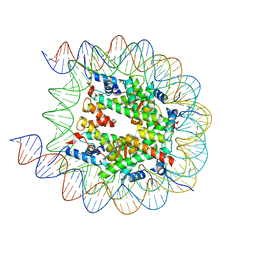 | | Cryo-EM structure of the human nucleosome with scFv | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLD
 
 | | Cryo-EM structure of the 145 bp human nucleosome containing acetylated H3 tail | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLA
 
 | | Cryo-EM structure of the human nucleosome lacking N-terminal region of H2A, H2B, H3, and H4 | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
7YML
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodobacter capsulatus | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Satoh, I, Kobayashi, Y, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Mizoguchi, A, Humbel, B.M, Madigan, M.T, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Rhodobacter capsulatus forms a compact crescent-shaped LH1-RC photocomplex.
Nat Commun, 14, 2023
|
|
8PKO
 
 | | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Hitchman, C.J, Lia, A, Bayo, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer).
To Be Published
|
|
5B6Q
 
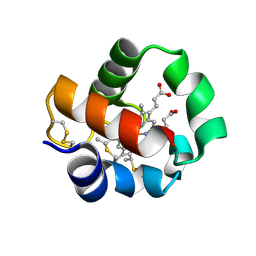 | | Crystal structure of monomeric cytochrome c5 from Shewanella violacea | | Descriptor: | HEME C, IMIDAZOLE, Soluble cytochrome cA | | Authors: | Masanari, M, Fujii, S, Kawahara, K, Oki, H, Tsujino, H, Maruno, T, Kobayashi, Y, Ohkubo, T, Nishiyama, M, Harada, Y, Wakai, S, Sambongi, Y. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-19 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Comparative study on stabilization mechanism of monomeric cytochrome c5 from deep-sea piezophilic Shewanella violacea
Biosci.Biotechnol.Biochem., 2016
|
|
3FHP
 
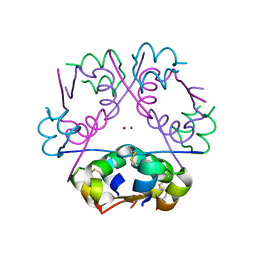 | | A neutron crystallographic analysis of a porcine 2Zn insulin at 2.0 A resolution | | Descriptor: | Insulin, ZINC ION | | Authors: | Iwai, W, Kurihara, K, Yamada, T, Kobayashi, Y, Ohnishi, Y, Tanaka, I, Takahashi, H, Niimura, N. | | Deposit date: | 2008-12-09 | | Release date: | 2009-10-20 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | A neutron crystallographic analysis of T6 porcine insulin at 2.1 A resolution
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5Z09
 
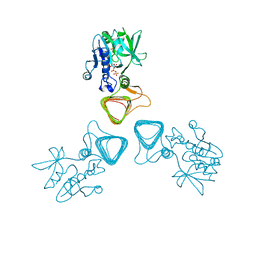 | | ST0452(Y97N)-UTP binding form | | Descriptor: | Dual sugar-1-phosphate nucleotidylyltransferase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Honda, Y, Nakano, S, Ito, S, Dadashipour, M, Zhang, Z, Kawarabayasi, Y. | | Deposit date: | 2017-12-19 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Improvement of ST0452N-Acetylglucosamine-1-Phosphate Uridyltransferase Activity by the Cooperative Effect of Two Single Mutations Identified through Structure-Based Protein Engineering
Appl. Environ. Microbiol., 84, 2018
|
|
5B3I
 
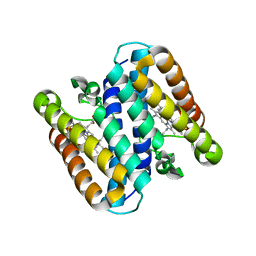 | | Homo-dimeric structure of cytochrome c' from Thermophilic Hydrogenophilus thermoluteolus | | Descriptor: | Cytochrome c prime, HEME C | | Authors: | Fujii, S, Oki, H, Kawahara, K, Yamane, D, Yamanaka, M, Maruno, T, Kobayashi, Y, Masanari, M, Wakai, S, Nishihara, H, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional insights into thermally stable cytochrome c' from a thermophile
Protein Sci., 26, 2017
|
|
6A3L
 
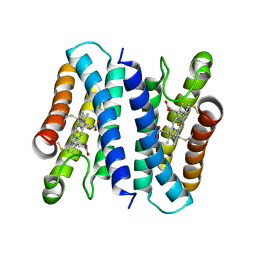 | | Crystal structure of cytochrome c' from Shewanella violacea DSS12 | | Descriptor: | Cytochrome c, HEME C | | Authors: | Suka, A, Oki, H, Kato, Y, Kawahara, K, Ohkubo, T, Maruno, T, Kobayashi, Y, Fujii, S, Wakai, S, Sambongi, Y. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-12 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Stability of cytochromes c' from psychrophilic and piezophilic Shewanella species: implications for complex multiple adaptation to low temperature and high hydrostatic pressure.
Extremophiles, 23, 2019
|
|
