3LUN
 
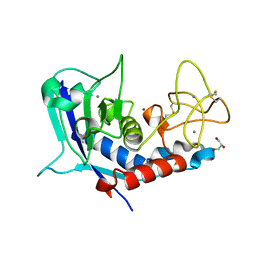 | | Structure of ulilysin mutant M290C | | Descriptor: | ARGININE, CALCIUM ION, GLYCEROL, ... | | Authors: | Tallant, C, Garcia-Castellanos, R, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the relevance of the Met-turn methionine in metzincins.
J.Biol.Chem., 285, 2010
|
|
2CEA
 
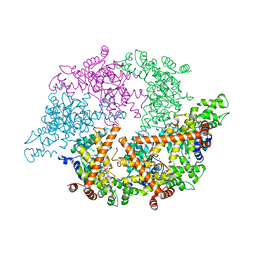 | | CELL DIVISION PROTEIN FTSH | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CELL DIVISION PROTEIN FTSH, MAGNESIUM ION, ... | | Authors: | Bieniossek, C, Baumann, U. | | Deposit date: | 2006-02-03 | | Release date: | 2006-02-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The Molecular Architecture of the Metalloprotease Ftsh.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
8PZ3
 
 | |
8Q00
 
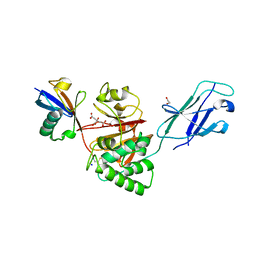 | | TssM-Ub-PA complex - A USP-like DUB from B. pseudomallei (193-430) reacted with Ub-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Polyubiquitin-B, ... | | Authors: | Uthoff, M, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structural basis for deubiquitination by the fingerless USP-type effector TssM.
Life Sci Alliance, 7, 2024
|
|
6EI1
 
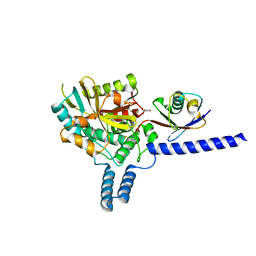 | | Crystal structure of the covalent complex between deubiquitinase ZUFSP (ZUP1) and Ubiquitin-PA | | Descriptor: | GLYCEROL, MALONATE ION, Polyubiquitin-B, ... | | Authors: | Pichlo, C, Baumann, U, Hofmann, K, Hermanns, T. | | Deposit date: | 2017-09-16 | | Release date: | 2018-03-07 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.732 Å) | | Cite: | A family of unconventional deubiquitinases with modular chain specificity determinants.
Nat Commun, 9, 2018
|
|
3CT4
 
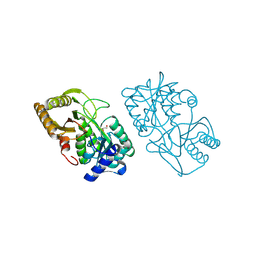 | | Structure of Dha-kinase subunit DhaK from L. Lactis | | Descriptor: | Dihydroxyacetone, PTS-dependent dihydroxyacetone kinase, dihydroxyacetone-binding subunit dhaK | | Authors: | Jeckelmann, J.M, Zurbriggen, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2008-04-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | X-ray Structures of the Three Lactococcus lactis Dihydroxyacetone Kinase Subunits and of a Transient Intersubunit Complex.
J.Biol.Chem., 283, 2008
|
|
3CR3
 
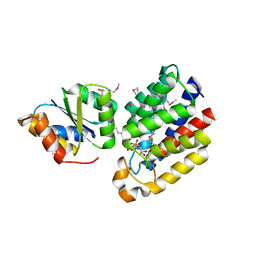 | | Structure of a transient complex between Dha-kinase subunits DhaM and DhaL from Lactococcus lactis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PTS-dependent dihydroxyacetone kinase, ... | | Authors: | Jeckelmann, J.M, Zurbriggen, A, Christen, S, Baumann, U, Erni, B. | | Deposit date: | 2008-04-04 | | Release date: | 2008-10-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray Structures of the Three Lactococcus lactis Dihydroxyacetone Kinase Subunits and of a Transient Intersubunit Complex.
J.Biol.Chem., 283, 2008
|
|
2J83
 
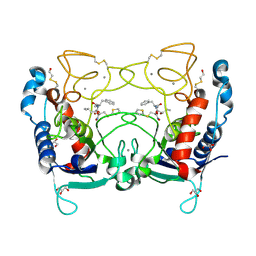 | | Ulilysin metalloprotease in complex with batimastat. | | Descriptor: | 4-(N-HYDROXYAMINO)-2R-ISOBUTYL-2S-(2-THIENYLTHIOMETHYL)SUCCINYL-L-PHENYLALANINE-N-METHYLAMIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Garcia-Castellanos, R, Tallant, C, Marrero, A, Sola, M, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2006-10-18 | | Release date: | 2006-12-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Specificity of a Metalloprotease of the Pappalysin Family Revealed by an Inhibitor and a Product Complex.
Arch.Biochem.Biophys., 457, 2007
|
|
1H5Q
 
 | | Mannitol dehydrogenase from Agaricus bisporus | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT MANNITOL DEHYDROGENASE, NICKEL (II) ION | | Authors: | Horer, S, Stoop, J, Mooibroek, H, Baumann, U, Sassoon, J. | | Deposit date: | 2001-05-24 | | Release date: | 2001-06-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Crystallographic Structure of the Mannitol 2-Dehydrogenase Nadp+ Binary Complex from Agaricus Bisporus
J.Biol.Chem., 276, 2001
|
|
6ZMK
 
 | | Crystal structure of human GFAT-1 L405R | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Mayr, F, Miethe, S, Atanassov, I, Baumann, U, Denzel, M.S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
6ZMJ
 
 | | Crystal structure of human GFAT-1 R203H | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Mayr, F, Miethe, S, Atanassov, I, Baumann, U, Denzel, M.S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.774 Å) | | Cite: | Protein kinase A controls the hexosamine pathway by tuning the feedback inhibition of GFAT-1.
Nat Commun, 12, 2021
|
|
7NZY
 
 | | Crystal structure of human Casein Kinase I delta in complex with CGS-15943 | | Descriptor: | 9-chloranyl-2-(furan-2-yl)-[1,2,4]triazolo[1,5-c]quinazolin-5-amine, Casein kinase I isoform delta, MALONATE ION, ... | | Authors: | Pichlo, C, Baumann, U. | | Deposit date: | 2021-03-24 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Phenotypic Discovery of Triazolo[1,5- c ]quinazolines as a First-In-Class Bone Morphogenetic Protein Amplifier Chemotype.
J.Med.Chem., 65, 2022
|
|
8ADD
 
 | | Viral tegument-like DUBs | | Descriptor: | ATP-dependent DNA helicase | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
3ZXC
 
 | |
6SVQ
 
 | | Crystal structure of human GFAT-1 G461E after UDP-GlcNAc soaking | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate-aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.717 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
3KDS
 
 | | apo-FtsH crystal structure | | Descriptor: | Cell division protein FtsH, N-{(2R)-2-[2-(hydroxyamino)-2-oxoethyl]-4-methylpentanoyl}-3-naphthalen-2-yl-L-alanyl-L-alaninamide, ZINC ION | | Authors: | Bieniossek, C, Niederhauser, B, Baumann, U. | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The crystal structure of apo-FtsH reveals domain movements necessary for substrate unfolding and translocation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6SVO
 
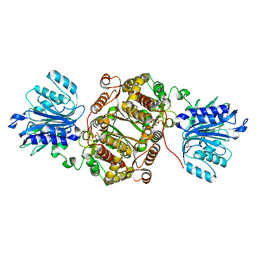 | | Crystal structure of human GFAT-1 in complex with Glucosamine-6-Phosphate and L-Glu | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1 | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
6SVM
 
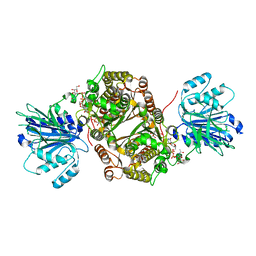 | | Crystal structure of human GFAT-1 in complex with Glucose-6-Phosphate, L-Glu, and UDP-GalNAc | | Descriptor: | GLUCOSE-6-PHOSPHATE, GLUTAMIC ACID, Glutamine--fructose-6-phosphate aminotransferase [isomerizing] 1, ... | | Authors: | Ruegenberg, S, Horn, M, Pichlo, C, Allmeroth, K, Baumann, U, Denzel, M.S. | | Deposit date: | 2019-09-18 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.481 Å) | | Cite: | Loss of GFAT-1 feedback regulation activates the hexosamine pathway that modulates protein homeostasis.
Nat Commun, 11, 2020
|
|
3ZXB
 
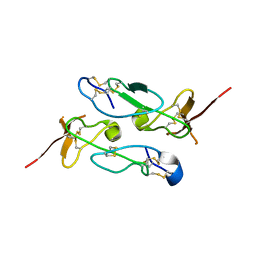 | |
4WW0
 
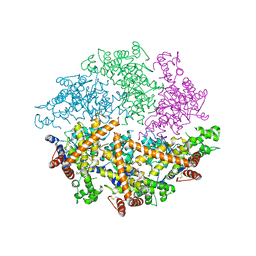 | | Truncated FtsH from A. aeolicus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH, ZINC ION | | Authors: | Vostrukhina, M, Baumann, U, Schacherl, M, Bieniossek, C, Lanz, M, Baumgartner, R. | | Deposit date: | 2014-11-09 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The structure of Aquifex aeolicus FtsH in the ADP-bound state reveals a C2-symmetric hexamer.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8OXI
 
 | |
8OXH
 
 | |
8OXL
 
 | |
8PHY
 
 | |
8OXK
 
 | |
