7SNY
 
 | | 2.10A Resolution Structure of NanoBiT Complementation Reporter Large Subunit LgBiT | | Descriptor: | Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.10A Resolution Structure of NanoBiT Complementation Reporter Large Subunit LgBiT
To be published
|
|
7SNT
 
 | | 2.20A Resolution Structure of NanoLuc Luciferase with Bound Substrate Analog 3-methoxy-furimazine | | Descriptor: | (4S)-8-benzyl-2-[(furan-2-yl)methyl]-3-methoxy-6-phenylimidazo[1,2-a]pyrazine, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Unch, J, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.20A Resolution Structure of NanoLuc Luciferase with Bound Substrate Analog 3-methoxy-furimazine
To be published
|
|
7SNR
 
 | | 2.00A Resolution Structure of NanoLuc Luciferase | | Descriptor: | ACETATE ION, CITRATE ANION, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2021-10-28 | | Release date: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00A Resolution Structure of NanoLuc Luciferase
To be published
|
|
7LKT
 
 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | Descriptor: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
4O8I
 
 | | 1.45A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, mitochondrial | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4O8H
 
 | | 0.85A resolution structure of PEG 400 Bound Cyclophilin D | | Descriptor: | DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, Peptidyl-prolyl cis-trans isomerase F, ... | | Authors: | Lovell, S, Valasani, K.R, Battaile, K.P, Wang, C, Yan, S.S. | | Deposit date: | 2013-12-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | High-resolution crystal structures of two crystal forms of human cyclophilin D in complex with PEG 400 molecules.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
7KNG
 
 | | 2.10A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-2 Y7F) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, DTY-ASP-TYR-PRO-GLY-ASP-PHE-CYS-TYR-LEU-TYR-GLY-THR-CYS, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
7KNF
 
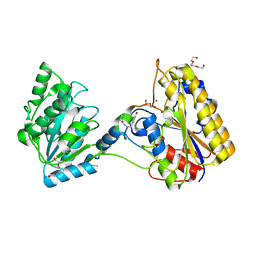 | | 1.80A resolution structure of independent Phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (Ce-1 NHOH) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, DTY-ASP-TYR-PRO-GLY-ASP-HIS-CYS-TYR-LEU-TYR-GLY-THR, SODIUM ION, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Weidmann, M, Dranchak, P, Aitha, M, Queme, B, Collmus, C.D, Kanter, L, Lamy, L, Tao, D, Rai, G, Suga, H, Inglese, J. | | Deposit date: | 2020-11-04 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-activity relationship of ipglycermide binding to phosphoglycerate mutases.
J.Biol.Chem., 296, 2021
|
|
4PEO
 
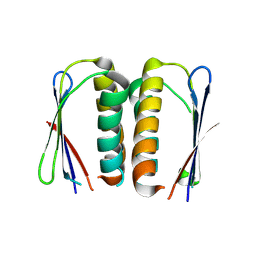 | | Crystal structure of a hypothetical protein from Staphylococcus aureus. | | Descriptor: | Hypothetical protein | | Authors: | McGrath, T.E, Kisselman, G, Romanov, V, Wu-Brown, J, Soloveychik, M, Chan, T.S.Y, Gordon, R.D, Thambipillai, D, Dharamsi, A, Mansoury, K, Battaile, K.P, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus.
To Be Published
|
|
5KGM
 
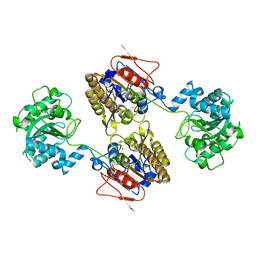 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
4QAS
 
 | | 1.27 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAQ
 
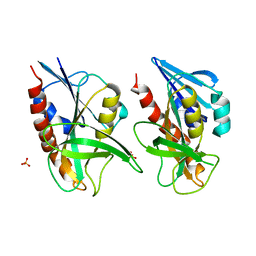 | | 1.58 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QFB
 
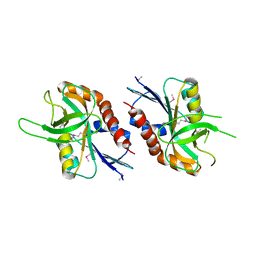 | | 1.99 A resolution structure of SeMet-CT263 (MTAN) from Chlamydia trachomatis | | Descriptor: | CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-20 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAT
 
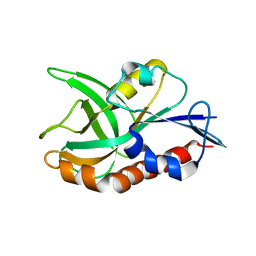 | | 1.75 A resolution structure of CT263-D161N (MTAN) from Chlamydia trachomatis bound to MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CT263 | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
4QAR
 
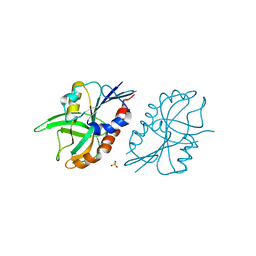 | | 1.45 A resolution structure of CT263 (MTAN) from Chlamydia trachomatis bound to Adenine | | Descriptor: | ADENINE, CT263, SULFATE ION | | Authors: | Barta, M.L, Thomas, K, Lovell, S, Battaile, K.P, Schramm, V.L, Hefty, P.S. | | Deposit date: | 2014-05-05 | | Release date: | 2014-10-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Biochemical Characterization of Chlamydia trachomatis Hypothetical Protein CT263 Supports That Menaquinone Synthesis Occurs through the Futalosine Pathway.
J.Biol.Chem., 289, 2014
|
|
7LKX
 
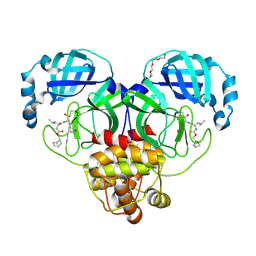 | | 1.60 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3e | | Descriptor: | (1R,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1S,2S,4S)-bicyclo[2.2.1]hept-5-en-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKS
 
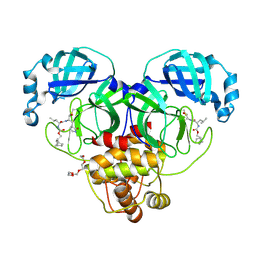 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2f | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKV
 
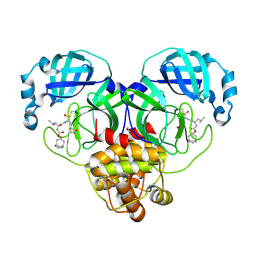 | | 1.55 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKR
 
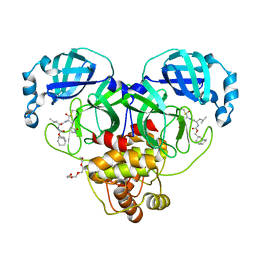 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2a | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKW
 
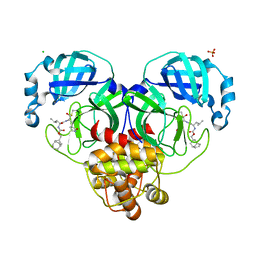 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3d (deuterated analog of inhibitor 3c) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKU
 
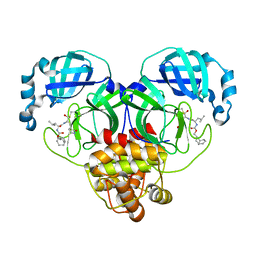 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3b (deuterated analog of inhibitor 2a) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
4PML
 
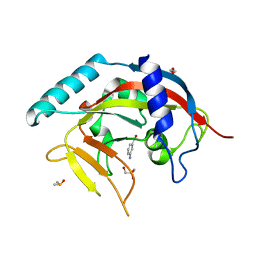 | | Crystal Structure of human Tankyrase 2 in complex with 3-amino-benzamide. | | Descriptor: | 1,2-ETHANEDIOL, 3-aminobenzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-22 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PNL
 
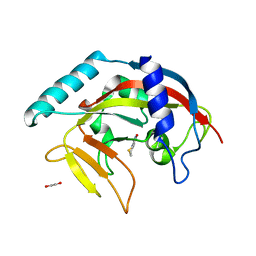 | | Crystal structure of TNKS-2 in complex with DR2313. | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,7,8-tetrahydro-4H-thiopyrano[4,3-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O6S
 
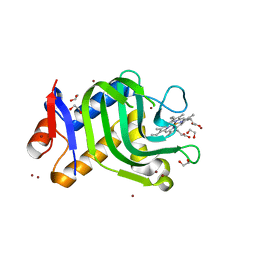 | | 1.32A resolution structure of the hemophore HasA from Pseudomonas aeruginosa (H83A mutant, Zinc Bound) | | Descriptor: | 1,2-ETHANEDIOL, HasAp, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Kumar, R, Battaile, K.P, Matsumura, H, Yao, H, Rodriguez, J.C, Moenne-Loccoz, P, Rivera, M. | | Deposit date: | 2013-12-23 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Replacing the Axial Ligand Tyrosine 75 or Its Hydrogen Bond Partner Histidine 83 Minimally Affects Hemin Acquisition by the Hemophore HasAp from Pseudomonas aeruginosa.
Biochemistry, 53, 2014
|
|
4PNS
 
 | | Crystal Structure of human Tankyrase 2 in complex with INH2BP. | | Descriptor: | 6-amino-5-iodo-2H-chromen-2-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
