8OWX
 
 | | Crystal Structure of METTL6 bound to SAH | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, S-ADENOSYL-L-HOMOCYSTEINE, tRNA N(3)-methylcytidine methyltransferase METTL6 | | Authors: | Throll, P, Basu, S, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-04-28 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 2024
|
|
8OWY
 
 | |
6S19
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from all available diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.Ch. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1D
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 4.57 keV from 20,000 diffraction patterns | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S1G
 
 | | Structure of thaumatin determined at SwissFEL using native-SAD at 6.06 keV from 50,000 diffraction patterns. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6S0Q
 
 | | Structure of the A2A adenosine receptor determined at SwissFEL using native-SAD at 4.57 keV from 50,000 diffraction patterns | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Nass, K, Cheng, R, Vera, L, Mozzanica, A, Redford, S, Ozerov, D, Basu, S, James, D, Knopp, G, Cirelli, C, Martiel, I, Casadei, C, Weinert, T, Nogly, P, Skopintsev, P, Usov, I, Leonarski, F, Geng, T, Rappas, M, Dore, A.S, Cooke, R, Nasrollahi Shirazi, S, Dworkowski, F, Sharpe, M, Olieric, N, Steinmetz, M.O, Schertler, G, Abela, R, Patthey, L, Schmitt, B, Hennig, M, Standfuss, J, Wang, M, Milne, J.C. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Advances in long-wavelength native phasing at X-ray free-electron lasers.
Iucrj, 7, 2020
|
|
6YX9
 
 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) at 2.4 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-04-30 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXG
 
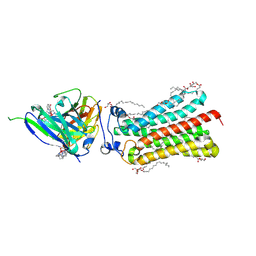 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Tb-XO4 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, OLEIC ACID, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXD
 
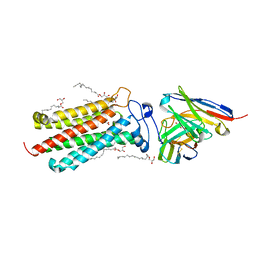 | | Room temperature structure of human adiponectin receptor 2 (ADIPOR2) at 2.9 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 2, GLYCEROL, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXF
 
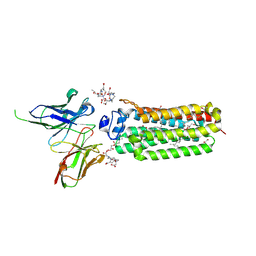 | | Cryogenic human adiponectin receptor 2 (ADIPOR2) with Gd-DO3 ligand determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Adiponectin receptor protein 2, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.025 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6YXH
 
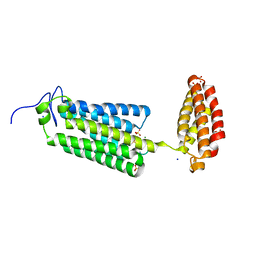 | | Cryogenic human alkaline ceramidase 3 (ACER3) at 2.6 A resolution determined by Serial Crystallography (SSX) using CrystalDirect | | Descriptor: | Alkaline ceramidase 3, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Healey, R.D, Basu, S, Humm, A.S, Leyrat, C, Dupeux, F, Pica, A, Granier, S, Marquez, J.A. | | Deposit date: | 2020-05-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An automated platform for structural analysis of membrane proteins through serial crystallography.
Cell Rep Methods, 1, 2021
|
|
6RPG
 
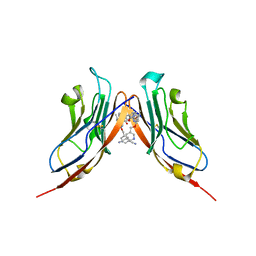 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with inhibitor | | Descriptor: | Programmed cell death 1 ligand 1, ~{N}-[2-[[4-[[3-[3-[[4-[(2-acetamidoethylamino)methyl]-5-[(5-cyanopyridin-3-yl)methoxy]-2-methyl-phenoxy]methyl]-2-methyl-phenyl]-2-methyl-phenyl]methoxy]-2-[(5-cyanopyridin-3-yl)methoxy]-5-methyl-phenyl]methylamino]ethyl]ethanamide | | Authors: | Magiera-Mularz, K, Basu, S, Yang, J, Xu, B, Skalniak, L, Musielak, B, Kholodovych, V, Holak, T.A, Hu, L. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design, Synthesis, Evaluation, and Structural Studies ofC2-Symmetric Small Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6FXM
 
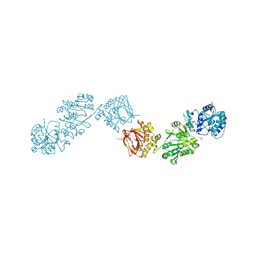 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Mn2+ | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6FXX
 
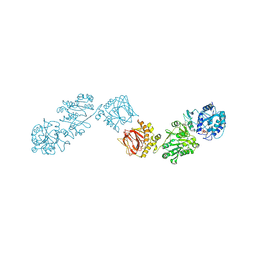 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Gal, Hg2+ Soak | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6FXY
 
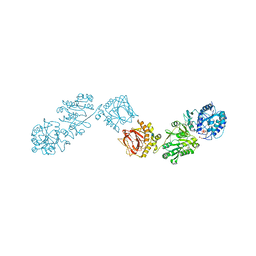 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Gal - Structure from long-wavelength S-SAD | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.138 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6FXK
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6FXT
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Glc | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6FXR
 
 | | Crystal Structure of full-length Human Lysyl Hydroxylase LH3 - Cocrystal with Fe2+, Mn2+, UDP-Gal | | Descriptor: | 2-OXOGLUTARIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scietti, L, Chiapparino, A, De Giorgi, F, Fumagalli, M, Khoriauli, L, Nergadze, S, Basu, S, Olieric, V, Banushi, B, Giulotto, E, Gissen, P, Forneris, F. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular architecture of the multifunctional collagen lysyl hydroxylase and glycosyltransferase LH3.
Nat Commun, 9, 2018
|
|
6G7O
 
 | | Crystal structure of human alkaline ceramidase 3 (ACER3) at 2.7 Angstrom resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Alkaline ceramidase 3,Soluble cytochrome b562, CALCIUM ION, ... | | Authors: | Leyrat, C, Vasiliauskaite-Brooks, I, Healey, R.D, Sounier, R, Grison, C, Hoh, F, Basu, S, Granier, S. | | Deposit date: | 2018-04-06 | | Release date: | 2019-01-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a human intramembrane ceramidase explains enzymatic dysfunction found in leukodystrophy.
Nat Commun, 9, 2018
|
|
1NQP
 
 | | Crystal structure of Human hemoglobin E at 1.73 A resolution | | Descriptor: | CYANIDE ION, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Dasgupta, J, Sen, U, Choudhury, D, Dutta, P, Basu, S, Chakrabarti, A, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2003-01-22 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallization and preliminary X-ray structural Studies of Hemoglobin A2 and Hemoglobin E, isolated from the blood samples of Beta-thalassemic patients
Biochem.Biophys.Res.Commun., 303, 2004
|
|
6FMV
 
 | | IMISX-EP of Hg-BacA soaking SIRAS | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMX
 
 | | IMISX-EP of W-PgpB | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphatidylglycerophosphatase B, TUNGSTATE(VI)ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMT
 
 | | IMISX-EP of Hg-BacA Soaking SAD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMY
 
 | | IMISX-EP of S-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMS
 
 | | IMISX-EP of Se-LspA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Globomycin, Lipoprotein signal peptidase | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
