1YUU
 
 | | Solution structure of Calcium-S100A13 | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2K6Y
 
 | | Solution structures of apo form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2L4O
 
 | | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain | | Descriptor: | Cell wall surface anchor family protein | | Authors: | Gentile, M.A, Melchiorre, S, Emolo, C, Moschioni, M, Gianfaldoni, C, Pancotto, L, Ferlenghi, I, Scarselli, M, Pansegrau, W, Veggi, D, Merola, M, Cantini, F, Ruggiero, P, Banci, L, Masignani, V. | | Deposit date: | 2010-10-11 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Streptococcus pneumoniae RrgB pilus backbone D1 domain: in vivo protection and implications for the molecular mechanisms of pilus polymerization
To be Published
|
|
1MFM
 
 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
1F22
 
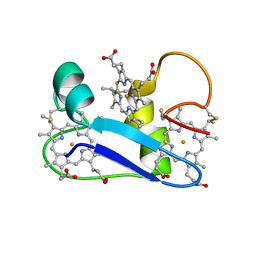 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS. COMPARISON BETWEEN THE REDUCED AND THE OXIDIZED FORMS. | | Descriptor: | CYTOCHROME C7, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | Deposit date: | 2000-05-23 | | Release date: | 2000-06-21 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
2NCL
 
 | |
1YS5
 
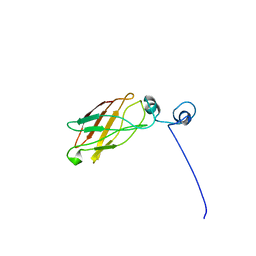 | | Solution structure of the antigenic domain of GNA1870 of Neisseria meningitidis | | Descriptor: | lipoprotein | | Authors: | Cantini, F, Savino, S, Masignani, V, Pizza, M, Scarselli, M, Swennen, E, Romagnoli, G, Veggi, D, Banci, L, Rappuoli, R. | | Deposit date: | 2005-02-07 | | Release date: | 2006-02-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant domain of protective antigen GNA1870 of Neisseria meningitidis
J.Biol.Chem., 281, 2006
|
|
1Z2G
 
 | | Solution structure of apo, oxidized yeast Cox17 | | Descriptor: | Cytochrome c oxidase copper chaperone | | Authors: | Arnesano, F, Balatri, E, Banci, L, Bertini, I, Winge, D.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-03-08 | | Release date: | 2005-06-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Folding studies of Cox17 reveal an important interplay of cysteine oxidation and copper binding
STRUCTURE, 13, 2005
|
|
2MPF
 
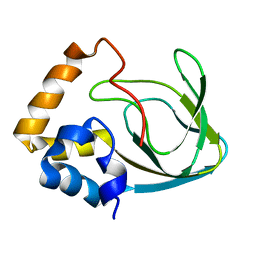 | | Solution structure human HCN2 CNBD in the cAMP-unbound state | | Descriptor: | Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Saponaro, A, Pauleta, S.R, Cantini, F, Matzapetakis, M, Hammann, C, Banci, L, Thiel, G, Santoro, B, Moroni, A. | | Deposit date: | 2014-05-16 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the mutual antagonism of cAMP and TRIP8b in regulating HCN channel function.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1NAQ
 
 | | Crystal structure of CUTA1 from E.coli at 1.7 A resolution | | Descriptor: | MERCURIBENZOIC ACID, MERCURY (II) ION, Periplasmic divalent cation tolerance protein cutA | | Authors: | Calderone, V, Mangani, S, Benvenuti, M, Viezzoli, M.S, Banci, L, Bertini, I, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-11-28 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction.
J.Biol.Chem., 278, 2003
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | Descriptor: | COPPER (II) ION, Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-21 | | Release date: | 2003-07-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
1ON4
 
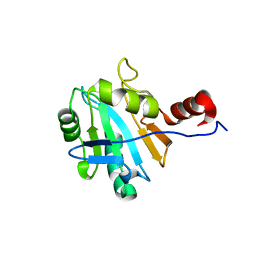 | | Solution structure of soluble domain of Sco1 from Bacillus Subtilis | | Descriptor: | Sco1 | | Authors: | Balatri, E, Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-02-27 | | Release date: | 2003-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Sco1: A Thioredoxin-like Protein Involved in Cytochrome c Oxidase Assembly
STRUCTURE, 11, 2003
|
|
2MJD
 
 | | Oxidized Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
1NM4
 
 | | Solution structure of Cu(I)-CopC from Pseudomonas syringae | | Descriptor: | Copper resistance protein C | | Authors: | Arnesano, F, Banci, L, Bertini, I, Mangani, S, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-01-09 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A redox switch in CopC: An intriguing copper trafficking protein that binds copper(I) and copper(II)
at different sites
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2LQL
 
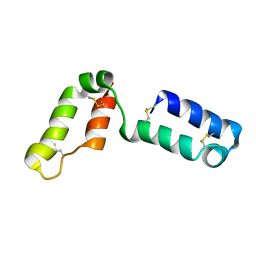 | | Solution structure of CHCH5 | | Descriptor: | Coiled-coil-helix-coiled-coil-helix domain-containing protein 5 | | Authors: | Peruzzini, R, Ciofi-Baffoni, S, Banci, L, Bertini, I. | | Deposit date: | 2012-03-09 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of CHCHD5 and CHCHD7: Two atypical human twin CX(9)C proteins.
J.Struct.Biol., 180, 2012
|
|
2JT5
 
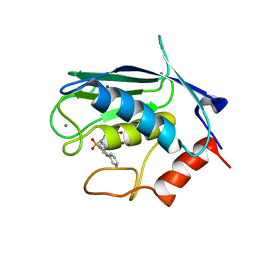 | | solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of n-hydroxy-2-[n-(2-hydroxyethyl)biphenyl-4-sulfonamide] hydroxamic acid (MLC88) | | Descriptor: | CALCIUM ION, N~2~-(biphenyl-4-ylsulfonyl)-N-hydroxy-N~2~-(2-hydroxyethyl)glycinamide, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-20 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2MJE
 
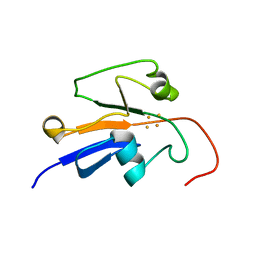 | | Reduced Yeast Adrenodoxin Homolog 1 | | Descriptor: | Adrenodoxin homolog, mitochondrial, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Gallo, A, Banci, L. | | Deposit date: | 2014-01-07 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional reconstitution of mitochondrial Fe/S cluster synthesis on Isu1 reveals the involvement of ferredoxin.
Nat Commun, 5, 2014
|
|
2JT6
 
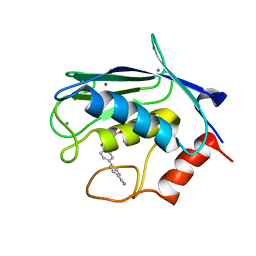 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of 3-4'-cyanobyphenyl-4-yloxy)-n-hdydroxypropionamide (MMP-3 inhibitor VII) | | Descriptor: | 3-[(4'-cyanobiphenyl-4-yl)oxy]-N-hydroxypropanamide, CALCIUM ION, Stromelysin-1, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-07-23 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2LQT
 
 | |
1M42
 
 | |
2JNP
 
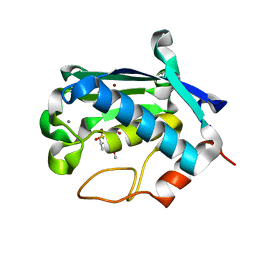 | | Solution structure of matrix metalloproteinase 3 (MMP-3) in the presence of N-isobutyl-N-[4-methoxyphenylsulfonyl]glycyl hydroxamic acid (NNGH) | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-3, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Alcaraz, L.A, Banci, L, Bertini, I, Cantini, F, Donaire, A, Gonnelli, L. | | Deposit date: | 2007-01-30 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Matrix metalloproteinase-inhibitor interaction: the solution structure of the catalytic domain of human matrix metalloproteinase-3 with different inhibitors
J.Biol.Inorg.Chem., 12, 2007
|
|
2JSD
 
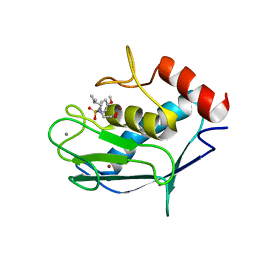 | | Solution structure of MMP20 complexed with NNGH | | Descriptor: | CALCIUM ION, Matrix metalloproteinase-20, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Arendt, Y, Banci, L, Bertini, I, Cantini, F, Cozzi, R, Del Conte, R, Gonnelli, L, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-07-03 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Catalytic domain of MMP20 (Enamelysin) - the NMR structure of a new matrix metalloproteinase.
Febs Lett., 581, 2007
|
|
2K6Z
 
 | | Solution structures of copper loaded form PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K70
 
 | | Solution structures of copper loaded form PCuA (cis conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | COPPER (I) ION, Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P.A, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
2K6W
 
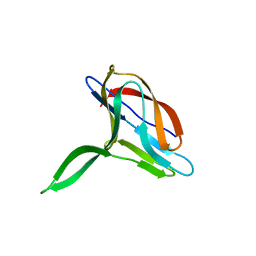 | | Solution structures of apo PCuA (trans conformation of the peptide bond involving the nitrogen of P14) | | Descriptor: | Putative uncharacterized protein TTHA1943 | | Authors: | Abriata, L.A, Banci, L, Bertini, I, Ciofi-Baffoni, S, Gkazonis, P, Spyroulias, G.A, Vila, A.J, Wang, S. | | Deposit date: | 2008-07-28 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mechanism of Cu(A) assembly.
Nat.Chem.Biol., 4, 2008
|
|
