6FWV
 
 | |
2VFF
 
 | | Crystal structure of the F96H mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-04 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1CGC
 
 | |
6G10
 
 | |
6G9V
 
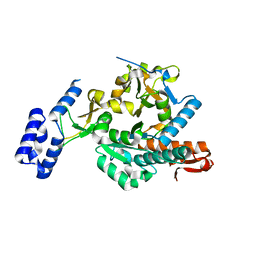 | | Crystal structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase(AfUAP1) in complex with UDPGlcNAc, pyrophosphate and Mg2+ | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE, UDP-N-acetylglucosamine pyrophosphorylase, ... | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
2VFE
 
 | | Crystal structure of F96S mutant of Plasmodium falciparum triosephosphate isomerase with 3- phosphoglycerate bound at the dimer interface | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, GLYCEROL, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
6FUB
 
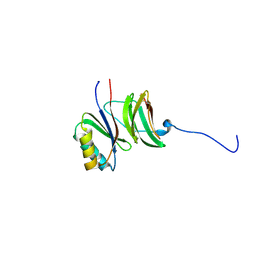 | |
6G9W
 
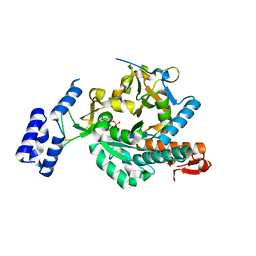 | | Crystal Structure of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase in complex with UTP | | Descriptor: | UDP-N-acetylglucosamine pyrophosphorylase, URIDINE 5'-TRIPHOSPHATE | | Authors: | Raimi, O.G, Hurtado-Guerrero, R, Borodin, V, Urbaniak, M, Ferguson, M, van Aalten, D. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A mechanism-inspired UDP-N-acetylglucosamine pyrophosphorylase inhibitor
RSC Chem. Biol., 2020
|
|
1N1Z
 
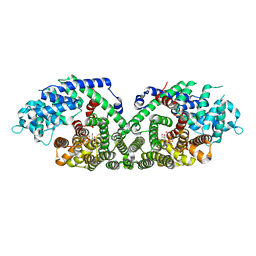 | | (+)-Bornyl Diphosphate Synthase: Complex with Mg and pyrophosphate | | Descriptor: | (+)-bornyl diphosphate synthase, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Whittington, D.A, Wise, M.L, Urbansky, M, Coates, R.M, Croteau, R.B, Christianson, D.W. | | Deposit date: | 2002-10-21 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6G11
 
 | |
1WKO
 
 | |
1R3O
 
 | | Crystal structure of the first RNA duplex in L-conformation at 1.9A resolution | | Descriptor: | L-RNA | | Authors: | Vallazza, M, Perbandt, M, Klussmann, S, Rypniewski, W, Erdmann, V.A, Betzel, C. | | Deposit date: | 2003-10-02 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | First look at RNA in L-configuration.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WKP
 
 | | Flowering locus t (ft) from arabidopsis thaliana | | Descriptor: | FLOWERING LOCUS T protein, SULFATE ION | | Authors: | Miller, D, Banfield, M.J, Winter, V.J, Brady, R.L. | | Deposit date: | 2004-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A divergent external loop confers antagonistic activity on floral regulators FT and TFL1.
Embo J., 25, 2006
|
|
2V7R
 
 | | Crystal structure of a human tRNAGly microhelix at 1.2 Angstrom resolution | | Descriptor: | HUMAN TRNAGLY MICROHELIX | | Authors: | Foerster, C, Mankowska, M, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-08-01 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a Human Trnagly Microhelix at 1.2 A Resolution.
Biochem.Biophys.Res.Commun., 368, 2008
|
|
2XV2
 
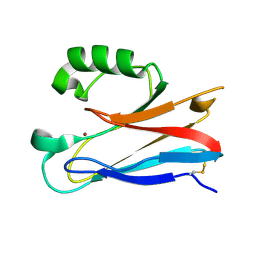 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.2 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
1E3S
 
 | | Rat brain 3-hydroxyacyl-CoA dehydrogenase binary complex with NADH | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SHORT CHAIN 3-HYDROXYACYL-COA DEHYDROGENASE | | Authors: | Powell, A.J, Read, J.A, Banfield, M.J, Brady, R.L. | | Deposit date: | 2000-06-22 | | Release date: | 2001-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of Structurally Diverse Substrates by Type II 3-Hydroxyacyl-Coa Dehydrogenase (Hadh II) Amyloid-Beta Binding Alcohol Dehydrogenase (Abad)
J.Mol.Biol., 303, 2000
|
|
3PVF
 
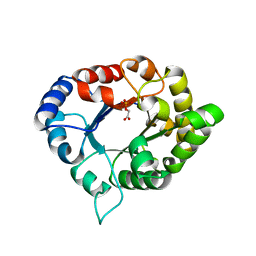 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase complexed with PGA | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-07 | | Release date: | 2011-04-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
3OFW
 
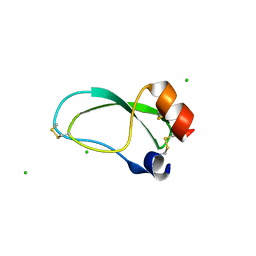 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Carribean sea anemone stichodactyla helianthus | | Descriptor: | CHLORIDE ION, Kunitz-type proteinase inhibitor SHPI-1 | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Talavera, A, Gil, D, Gonzalez, Y, de los Angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-08-16 | | Release date: | 2011-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the recombinant BPTI/Kunitz-type inhibitor rShPI-1A from the marine invertebrate Stichodactyla helianthus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
2XV0
 
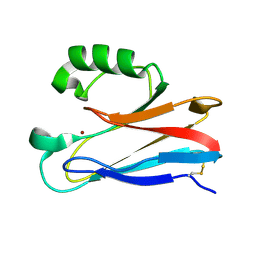 | | Pseudomonas aeruginosa Azurin with mutated metal-binding loop sequence (CAAHAAM), chemically reduced, pH4.8 | | Descriptor: | AZURIN, COPPER (I) ION | | Authors: | Li, C, Sato, K, Monari, S, Salard, I, Sola, M, Banfield, M.J, Dennison, C. | | Deposit date: | 2010-10-22 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal-Binding Loop Length is a Determinant of the Pka of a Histidine Ligand at a Type 1 Copper Site
Inorg.Chem., 50, 2011
|
|
3PWA
 
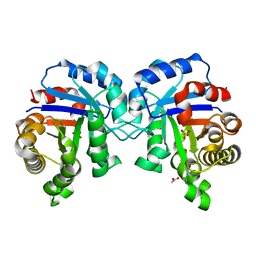 | | Structure of C126A mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, SULFATE ION, ... | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
2W89
 
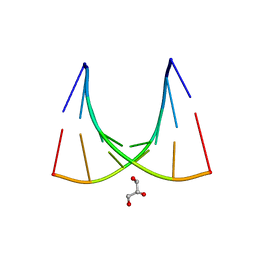 | | Crystal structure of the E.coli tRNAArg aminoacyl stem issoacceptor RR-1660 at 2.0 Angstroem resolution | | Descriptor: | 5'-R(*CP*GP*GP*AP*UP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*CP*CP*GP)-3', GLYCEROL | | Authors: | Eichert, A, Schreiber, A, Fuerste, J.P, Perbandt, M, Betzel, C, Erdmann, V.A, Foerster, C. | | Deposit date: | 2009-01-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the E. Coli tRNA(Arg) Aminoacyl Stem Isoacceptor Rr-1660 at 2.0 A Resolution.
Biochem.Biophys.Res.Commun., 385, 2009
|
|
2VAL
 
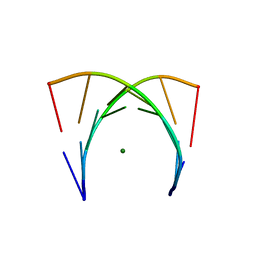 | | Crystal structure of an Escherichia coli tRNAGly microhelix at 2.0 Angstrom resolution | | Descriptor: | 5'-R(*GP*CP*GP*GP*GP*AP*AP)-3', 5'-R(*UP*UP*CP*CP*CP*GP*CP)-3', MAGNESIUM ION | | Authors: | Forster, C, Brauer, A.B.E, Perbandt, M, Lehmann, D, Furste, J.P, Betzel, C, Erdmann, V.A. | | Deposit date: | 2007-09-03 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of an Escherichia Coli Trnagly Microhelix at 2.0 Angstrom Resolution
Biochem.Biophys.Res.Commun., 363, 2007
|
|
2ARM
 
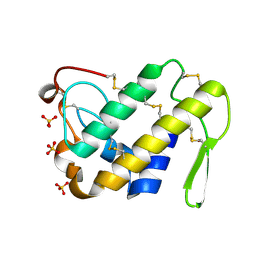 | | Crystal Structure of the Complex of Phospholipase A2 with a natural compound atropine at 1.2 A resolution | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | Authors: | Singh, N, Pal, A, Jabeen, T, Sharma, S, Perbandt, M, Betzel, C, Singh, T.P. | | Deposit date: | 2005-08-20 | | Release date: | 2005-09-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystal structures of the complexes of a group IIA phospholipase A2 with two natural anti-inflammatory agents, anisic acid, and atropine reveal a similar mode of binding
Proteins, 64, 2006
|
|
3PY2
 
 | | Structure of C126S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Triosephosphate isomerase | | Authors: | Samanta, M, Banerjee, M, Murthy, M.R.N, Balaram, H, Balaram, P. | | Deposit date: | 2010-12-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Probing the role of the fully conserved Cys126 in triosephosphate isomerase by site-specific mutagenesis--distal effects on dimer stability.
Febs J., 278, 2011
|
|
1LDG
 
 | | PLASMODIUM FALCIPARUM L-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C, Banfield, M, Barker, J, Higham, C, Moreton, K, Turgut-Balik, D, Brady, L, Holbrook, J.J. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of lactate dehydrogenase from Plasmodium falciparum reveals a new target for anti-malarial design.
Nat.Struct.Biol., 3, 1996
|
|
