5WMV
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WMX
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.M. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
6UD5
 
 | | Crystal structure of human tryptophan 2,3-dioxygenase in complex with carbon monoxide and tryptophan | | Descriptor: | CARBON MONOXIDE, PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, ... | | Authors: | Pham, K.N, Lewis-Ballester, A, Yeh, S.R. | | Deposit date: | 2019-09-18 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Plasticity in Human Heme-Based Dioxygenases.
J.Am.Chem.Soc., 143, 2021
|
|
5WMW
 
 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WMU
 
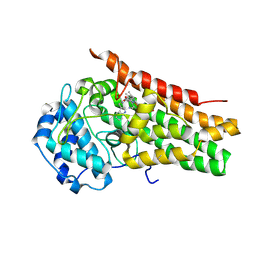 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase I | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.R, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
5WN8
 
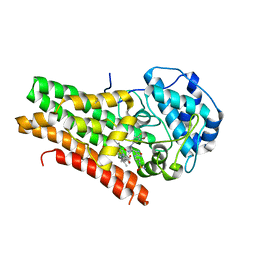 | | Structural Insights into Substrate and Inhibitor Binding Sites in Human Indoleamine 2,3-Dioxygenase 1 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(3-bromo-4-fluorophenyl)-N'-hydroxy-4-{[2-(sulfamoylamino)ethyl]amino}-1,2,5-oxadiazole-3-carboximidamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lewis-Ballester, A, Pham, K.N, Batabyal, D, Karkashon, S, Bonanno, J.B, Poulos, T.L, Yeh, S.R. | | Deposit date: | 2017-07-31 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into substrate and inhibitor binding sites in human indoleamine 2,3-dioxygenase 1.
Nat Commun, 8, 2017
|
|
6CXV
 
 | | Structure of the S167H mutant of human indoleamine 2,3 dioxygenase in complex with tryptophan and cyanide | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Lewis-Ballester, A, Yeh, S.-R, Karkashon, S, Batabyal, D, Poulos, T.L. | | Deposit date: | 2018-04-04 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
6CXU
 
 | | Structure of the S167H mutant of human indoleamine 2,3 dioxygenase in complex with tryptophan and cyanide | | Descriptor: | CYANIDE ION, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lewis-Ballester, A, Yeh, S.-R, Karkashon, S, Batabyal, D, Poulos, T.L. | | Deposit date: | 2018-04-04 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Inhibition Mechanisms of Human Indoleamine 2,3 Dioxygenase 1.
J. Am. Chem. Soc., 140, 2018
|
|
6E35
 
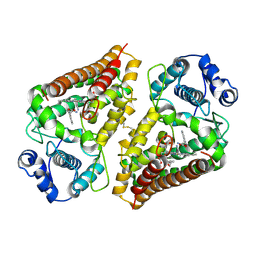 | | Crystal structure of human indoleamime 2,3-dioxygenase (IDO1) in complex with L-Trp and cyanide, Northeast Structural Genomics Target HR6160 | | Descriptor: | 2-(1H-indol-3-yl)ethanol, CYANIDE ION, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Forouhar, F, Lewis-Ballester, A, Lew, S, Karkashon, S, Seetharaman, J, Lu, C, Hussain, M, Yeh, S.-R, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-07-13 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.411 Å) | | Cite: | Crystal structure of human indoleamime 2,3-dioxygenase (IDO1) in complex with L-Trp and cyanide, Northeast Structural Genomics Target HR6160
To Be Published
|
|
6PZ1
 
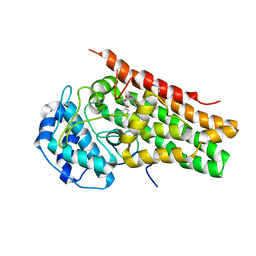 | | Crystal Structure of human Indoleamine 2,3-Dioxygenase 1 in complex with PF-06840003 in Active Site and Si site | | Descriptor: | (3R)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Pham, K.N, Lewis-Ballester, A, Yeh, S.R. | | Deposit date: | 2019-07-31 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in Human Indoleamine 2,3-Dioxygenase 1 and Tryptophan Dioxygenase.
J.Am.Chem.Soc., 141, 2019
|
|
6PYZ
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PF-06840003 in Active Site | | Descriptor: | (3S)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase, ... | | Authors: | Pham, K.N, Lewis-Ballester, A, Yeh, S.R. | | Deposit date: | 2019-07-31 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in Human Indoleamine 2,3-Dioxygenase 1 and Tryptophan Dioxygenase.
J.Am.Chem.Soc., 141, 2019
|
|
6PYY
 
 | | Crystal Structure of human Tryptophan 2,3-dioxygenase in complex with PF-06840003 in Active Site and Exo site | | Descriptor: | (3S)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, PROTOPORPHYRIN IX CONTAINING FE, Tryptophan 2,3-dioxygenase | | Authors: | Pham, K.N, Lewis-Ballester, A, Yeh, S.R. | | Deposit date: | 2019-07-31 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in Human Indoleamine 2,3-Dioxygenase 1 and Tryptophan Dioxygenase.
J.Am.Chem.Soc., 141, 2019
|
|
2CHZ
 
 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-93 | | Descriptor: | N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2CHX
 
 | | A pharmacological map of the PI3-K family defines a role for p110alpha in signaling: The structure of complex of phosphoinositide 3-kinase gamma with inhibitor PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
2CHW
 
 | | A pharmacological map of the PI3-K family defines a role for p110 alpha in signaling: The structure of complex of phosphoinositide 3- kinase gamma with inhibitor PIK-39 | | Descriptor: | 2-((9H-PURIN-6-YLTHIO)METHYL)-5-CHLORO-3-(2-METHOXYPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM | | Authors: | Knight, Z.A, Gonzalez, B, Feldman, M.E, Zunder, E.R, Goldenberg, D.D, Williams, O, Loewith, R, Stokoe, D, Balla, A, Toth, B, Balla, T, Weiss, W.A, Williams, R.L, Shokat, K.M. | | Deposit date: | 2006-03-16 | | Release date: | 2006-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Pharmacological Map of the Pi3-K Family Defines a Role for P110Alpha in Signaling
Cell(Cambridge,Mass.), 125, 2006
|
|
3M9G
 
 | | Crystal structure of the three-PASTA-domain of a Ser/Thr kinase from Staphylococcus aureus | | Descriptor: | Protein kinase, ZINC ION | | Authors: | Paracuellos, P, Ballandras, A, Robert, X, Creze, C, Cozzone, A.J, Duclos, B, Gouet, P. | | Deposit date: | 2010-03-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Extended Conformation of the 2.9-A Crystal Structure of the Three-PASTA Domain of a Ser/Thr Kinase from the Human Pathogen Staphylococcus aureus
J.Mol.Biol., 404, 2010
|
|
6EI6
 
 | | CC2D1B coordinates ESRCT-III activity during the mitotic reformation of the nuclear envelope | | Descriptor: | Coiled-coil and C2 domain-containing protein 1-like, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Ventimiglia, L.N, Cuesta-Geijo, M.A, Martinelli, N, Caballe, A, Macheboeuf, P, Miguet, N, Parnham, I.M, Olmos, Y, Carlton, J.G, Weissehorn, W, martin-Serrano, J. | | Deposit date: | 2017-09-18 | | Release date: | 2018-10-10 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.461 Å) | | Cite: | CC2D1B Coordinates ESCRT-III Activity during the Mitotic Reformation of the Nuclear Envelope.
Dev. Cell, 47, 2018
|
|
6F09
 
 | | Binary complex of 14-3-3 zeta with ubiquitin specific protease 8 (USP8) pSer718 peptide | | Descriptor: | 14-3-3 protein zeta/delta, Ubiquitin carboxyl-terminal hydrolase 8 | | Authors: | Centorrino, F, Ballone, A, Ottmann, C, Guo, S, Leysen, S. | | Deposit date: | 2017-11-17 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.594 Å) | | Cite: | Biophysical and structural insight into the USP8/14-3-3 interaction.
FEBS Lett., 592, 2018
|
|
4A5Z
 
 | | Structures of MITD1 | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-30 | | Release date: | 2012-11-14 | | Last modified: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AE4
 
 | | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a novel SOUBA domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Agromayor, M, Soler, N, Caballe, A, Kueck, T, Freund, S.M, Allen, M.D, Bycroft, M, Perisic, O, Ye, Y, McDonald, B, Scheel, H, Hofmann, K, Neil, S.J.D, Martin-Serrano, J, Williams, R.L. | | Deposit date: | 2012-01-06 | | Release date: | 2012-03-21 | | Last modified: | 2018-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The UBAP1 subunit of ESCRT-I interacts with ubiquitin via a SOUBA domain.
Structure, 20, 2012
|
|
4A5X
 
 | | Structures of MITD1 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, CHARGED MULTIVESICULAR BODY PROTEIN 1A, GLYCEROL, ... | | Authors: | Hadders, M.A, Agromayor, M, Caballe, A, Obita, T, Perisic, O, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2011-10-28 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3BIA
 
 | | Tim-4 in complex with sodium potassium tartrate | | Descriptor: | L(+)-TARTARIC ACID, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3BI9
 
 | | Tim-4 | | Descriptor: | ACETATE ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3BIB
 
 | | Tim-4 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
2OR8
 
 | | Tim-1 | | Descriptor: | ACETATE ION, Hepatitis A virus cellular receptor 1 homolog | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Casasnovas, J.M. | | Deposit date: | 2007-02-02 | | Release date: | 2007-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Receptors 1 and 2 Reveal Mechanisms for Regulation of Immune Responses by the TIM Receptor Family.
Immunity, 26, 2007
|
|
