3O2O
 
 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
1N6G
 
 | | The structure of immature Dengue-2 prM particles | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-10 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structures of Immature flavivirus particles
EMBO J., 22, 2003
|
|
3J0F
 
 | | Sindbis virion | | Descriptor: | Capsid protein, E1 envelope glycoprotein, E2 envelope glycoprotein | | Authors: | Tang, J, Jose, J, Zhang, W, Chipman, P, Kuhn, R.J, Baker, T.S. | | Deposit date: | 2011-07-08 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Molecular Links between the E2 Envelope Glycoprotein and Nucleocapsid Core in Sindbis Virus.
J.Mol.Biol., 414, 2011
|
|
1ZSZ
 
 | | Crystal structure of a computationally designed SspB heterodimer | | Descriptor: | MAGNESIUM ION, Stringent starvation protein B homolog | | Authors: | Bolon, D.N, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2005-05-25 | | Release date: | 2005-08-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity versus stability in computational protein design.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1YFN
 
 | | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB- the crystal structure of a SspB-RseA complex | | Descriptor: | Sigma-E factor negative regulatory protein, Stringent starvation protein B | | Authors: | Levchenko, I, Grant, R.A, Flynn, J.M, Sauer, R.T, Baker, T.A. | | Deposit date: | 2005-01-03 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile modes of peptide recognition by the AAA+ adaptor protein SspB
Nat.Struct.Mol.Biol., 12, 2005
|
|
1Z8Y
 
 | | Mapping the E2 Glycoprotein of Alphaviruses | | Descriptor: | Capsid protein C, Spike glycoprotein E1, Spike glycoprotein E2 | | Authors: | Mukhopadhyay, S, Zhang, W, Gabler, S, Chipman, P.R, Strauss, E.G, Strauss, J.H, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-03-31 | | Release date: | 2006-02-07 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Mapping the structure and function of the E1 and E2 glycoproteins in alphaviruses.
Structure, 14, 2006
|
|
1NA4
 
 | | The structure of immature Yellow Fever virus particle | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
7UIY
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIW
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UJ0
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIIb | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIZ
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIc | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIX
 
 | | ClpAP complex bound to ClpS N-terminal extension, class I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UIV
 
 | | ClpAP complex bound to ClpS N-terminal extension, class IIa | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent Clp protease ATP-binding subunit ClpA, ATP-dependent Clp protease adapter protein ClpS, ... | | Authors: | Kim, S, Fei, X, Sauer, R.T, Baker, T.A. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | AAA+ protease-adaptor structures reveal altered conformations and ring specialization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
3UX1
 
 | | Structural Characterization of Adeno-Associated Virus Serotype 9 | | Descriptor: | Capsid protein VP1 | | Authors: | DiMattia, M.A, Nam, H.-J, Van Vliet, K, Mitchell, M, McCall, A, Bennett, A, Gurda, B, McKenna, R, Potter, M, Sakai, Y, Byrne, B.J, Muzyczka, N, Aslanidi, G, Zolotukhin, S, Olson, N, Sinkovitis, R, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2011-12-03 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the unique properties of adeno-associated virus serotype 9.
J.Virol., 86, 2012
|
|
5TXT
 
 | | Structure of asymmetric apo/holo ALAS dimer from S. cerevisiae | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 5-aminolevulinate synthase, mitochondrial, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
5TXR
 
 | | Structure of ALAS from S. cerevisiae non-covalently bound to PLP cofactor | | Descriptor: | 5-aminolevulinate synthase, mitochondrial, FORMIC ACID, ... | | Authors: | Brown, B.L, Grant, R.A, Kardon, J.R, Sauer, R.T, Baker, T.A. | | Deposit date: | 2016-11-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Mitochondrial Aminolevulinic Acid Synthase, a Key Heme Biosynthetic Enzyme.
Structure, 26, 2018
|
|
5F72
 
 | | De novo design and crystallographic validation of antibodies targeting a pre-selected epitope | | Descriptor: | Kelch-like ECH-associated protein 1, Single chain Fv from a Fab | | Authors: | Liu, X, Taylor, R.D, Griffin, L, Coker, S, Adams, R, Ceska, T, Shi, J, Lawson, A.D.G, Baker, T. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo design and crystallographic validation of antibodies targeting a pre-selected epitope
To Be Published
|
|
7M1M
 
 | | Crystal structure of Pseudomonas aeruginosa ClpP1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Mawla, G.D, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
4YKA
 
 | | The structure of Agrobacterium tumefaciens ClpS2 in complex with L-tyrosinamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, L-TYROSINAMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
4YJX
 
 | | The structure of Agrobacterium tumefaciens ClpS2 bound to L-phenylalaninamide | | Descriptor: | ATP-dependent Clp protease adapter protein ClpS 2, PHENYLALANINE AMIDE, SULFATE ION | | Authors: | Stein, B, Grant, R.A, Sauer, R.T, Baker, T.A. | | Deposit date: | 2015-03-03 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Structural Basis of an N-Degron Adaptor with More Stringent Specificity.
Structure, 24, 2016
|
|
3ES5
 
 | | Crystal Structure of Partitivirus (PsV-F) | | Descriptor: | Putative capsid protein | | Authors: | Pan, J, Dong, L, Lin, L, Ochoa, W.F, Sinkovits, R.S, Havens, W.M, Nibert, M.L, Baker, T.S, Ghabrial, S.A, Tao, Y.J. | | Deposit date: | 2008-10-03 | | Release date: | 2009-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Atomic structure reveals the unique capsid organization of a dsRNA virus.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4YJM
 
 | |
1LD4
 
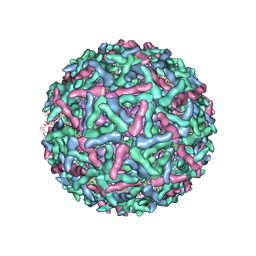 | | Placement of the Structural Proteins in Sindbis Virus | | Descriptor: | Coat protein C, GENERAL CONTROL PROTEIN GCN4, Spike glycoprotein E1, ... | | Authors: | Zhang, W, Mukhopadhyay, S, Pletnev, S.V, Baker, T.S, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-04-08 | | Release date: | 2002-11-04 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Placement of the Structural Proteins in Sindbis virus
J.VIROL., 76, 2002
|
|
7M1L
 
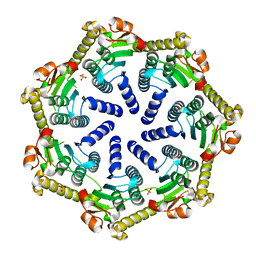 | | Crystal structure of Pseudomonas aeruginosa ClpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit, PHOSPHATE ION | | Authors: | Hall, B.M, Grant, R.A, Baker, T.A, Sauer, R.T. | | Deposit date: | 2021-03-13 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa.
Mol.Microbiol., 115, 2021
|
|
