3BLG
 
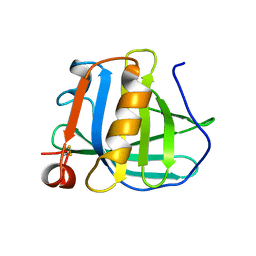 | | STRUCTURAL BASIS OF THE TANFORD TRANSITION OF BOVINE BETA-LACTOGLOBULIN FROM CRYSTAL STRUCTURES AT THREE PH VALUES; PH 6.2 | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Qin, B.Y, Bewley, M.C, Creamer, L.K, Baker, H.M, Baker, E.N, Jameson, G.B. | | Deposit date: | 1998-08-29 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis of the Tanford transition of bovine beta-lactoglobulin.
Biochemistry, 37, 1998
|
|
1TY2
 
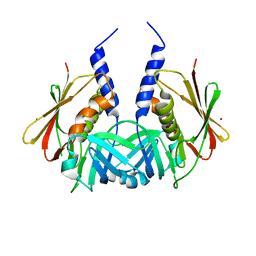 | | Crystal structure of the streptococcal pyrogenic exotoxin J (SPE-J) | | Descriptor: | ZINC ION, putative exotoxin (superantigen) | | Authors: | Baker, H.M, Proft, T, Webb, P.D, Arcus, V.L, Fraser, J.D, Baker, E.N. | | Deposit date: | 2004-07-07 | | Release date: | 2004-08-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and mutational data show that the streptococcal pyrogenic exotoxin j can use a common binding surface for T-cell receptor binding and dimerization
J.Biol.Chem., 279, 2004
|
|
1CB6
 
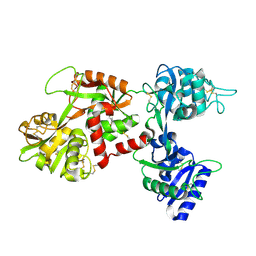 | | STRUCTURE OF HUMAN APOLACTOFERRIN AT 2.0 A RESOLUTION. | | Descriptor: | CHLORIDE ION, Lactotransferrin | | Authors: | Jameson, G.B, Anderson, B.F, Norris, G.E, Thomas, D.H, Baker, E.N. | | Deposit date: | 1999-03-01 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human apolactoferrin at 2.0 A resolution. Refinement and analysis of ligand-induced conformational change.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1TY0
 
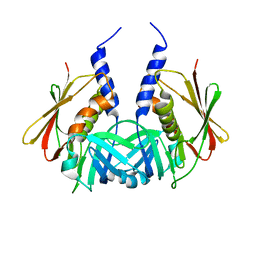 | | Crystal structure of the streptococcal pyrogenic exotoxin J (SPE-J) | | Descriptor: | putative exotoxin (superantigen) | | Authors: | Baker, H.M, Proft, T, Webb, P.D, Arcus, V.L, Fraser, J.D, Baker, E.N. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystallographic and mutational data show that the streptococcal pyrogenic exotoxin j can use a common binding surface for T-cell receptor binding and dimerization
J.Biol.Chem., 279, 2004
|
|
5FCC
 
 | | Structure of HutD from Pseudomonas fluorescens SBW25 (NaCl condition) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, HutD, ... | | Authors: | Johnston, J.M, Gerth, M.L, Baker, E.N, Lott, J.S, Rainey, P.B. | | Deposit date: | 2015-12-15 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure of HutD from Pseudomonas fluorescens
To Be Published
|
|
5E5Q
 
 | | Racemic snakin-1 in P21/c | | Descriptor: | Snakin-1 | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5E5Y
 
 | | Quasi-racemic snakin-1 in P1 before radiation damage | | Descriptor: | 1,2-ETHANEDIOL, D- snakin-1, FORMIC ACID, ... | | Authors: | Yeung, H, Squire, C.J, Yosaatmadja, Y, Panjikar, S, Baker, E.N, Harris, P.W.R, Brimble, M.A. | | Deposit date: | 2015-10-09 | | Release date: | 2016-05-18 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.506 Å) | | Cite: | Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1VEW
 
 | | MANGANESE SUPEROXIDE DISMUTASE FROM ESCHERICHIA COLI | | Descriptor: | HYDROXIDE ION, MANGANESE (II) ION, MANGANESE SUPEROXIDE DISMUTASE | | Authors: | Edwards, R.A, Baker, H.M, Whittaker, M.M, Whittaker, J.W, Jameson, G.B, Baker, E.N. | | Deposit date: | 1998-01-20 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Escherichia coli manganese superoxide dismutase at 2.1-angstrom resolution.
J.Biol.Inorg.Chem., 3, 1998
|
|
1BKA
 
 | | OXALATE-SUBSTITUTED DIFERRIC LACTOFERRIN | | Descriptor: | FE (III) ION, LACTOFERRIN, OXALATE ION | | Authors: | Baker, H.M, Smith, C.A, Baker, E.N. | | Deposit date: | 1996-04-15 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Anion binding by transferrins: importance of second-shell effects revealed by the crystal structure of oxalate-substituted diferric lactoferrin.
Biochemistry, 35, 1996
|
|
1ET6
 
 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 FROM STREPTOCOCCUS PYOGENES | | Descriptor: | SUPERANTIGEN SMEZ-2 | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1EU3
 
 | | CRYSTAL STRUCTURE OF THE SUPERANTIGEN SMEZ-2 (ZINC BOUND) FROM STREPTOCOCCUS PYOGENES | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, SUPERANTIGEN SMEZ-2, ... | | Authors: | Arcus, V.L, Proft, T, Sigrell, J.A, Baker, H.M, Fraser, J.D, Baker, E.N. | | Deposit date: | 2000-04-13 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Conservation and variation in superantigen structure and activity highlighted by the three-dimensional structures of two new superantigens from Streptococcus pyogenes.
J.Mol.Biol., 299, 2000
|
|
1EVJ
 
 | | CRYSTAL STRUCTURE OF GLUCOSE-FRUCTOSE OXIDOREDUCTASE (GFOR) DELTA1-22 S64D | | Descriptor: | GLUCOSE-FRUCTOSE OXIDOREDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lott, J.S, Halbig, D, Baker, H.M, Hardman, M.J, Sprenger, G.A, Baker, E.N. | | Deposit date: | 2000-04-20 | | Release date: | 2000-12-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a truncated mutant of glucose-fructose oxidoreductase shows that an N-terminal arm controls tetramer formation.
J.Mol.Biol., 304, 2000
|
|
1FQE
 
 | | CRYSTAL STRUCTURES OF MUTANT (K206A) THAT ABOLISH THE DILYSINE INTERACTION IN THE N-LOBE OF HUMAN TRANSFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, POTASSIUM ION, ... | | Authors: | Nurizzo, D, Baker, H.M, Baker, E.N. | | Deposit date: | 2000-09-04 | | Release date: | 2001-05-16 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and iron release properties of mutants (K206A and K296A) that abolish the dilysine interaction in the N-lobe of human transferrin.
Biochemistry, 40, 2001
|
|
1FQF
 
 | |
1LCT
 
 | |
6BBT
 
 | | Structure of the major pilin protein (T-13) from Streptococcus pyogenes serotype GAS131465 | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Young, P.G, Baker, E.N, Moreland, N.J. | | Deposit date: | 2017-10-19 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Group AStreptococcusT Antigens Have a Highly Conserved Structure Concealed under a Heterogeneous Surface That Has Implications for Vaccine Design.
Infect.Immun., 87, 2019
|
|
6BBW
 
 | |
4XOO
 
 | | FMN complex of coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain) | | Descriptor: | Coenzyme F420:L-glutamate ligase, FLAVIN MONONUCLEOTIDE | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
4XOQ
 
 | | F420 complex of coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain) | | Descriptor: | COENZYME F420, Coenzyme F420:L-glutamate ligase, SULFATE ION | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
4XOM
 
 | | Coenzyme F420:L-glutamate ligase (FbiB) from Mycobacterium tuberculosis (C-terminal domain). | | Descriptor: | Coenzyme F420:L-glutamate ligase, SULFATE ION | | Authors: | Rehan, A.M, Bashiri, G, Baker, H.M, Baker, E.N, Squire, C.J. | | Deposit date: | 2015-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elongation of the Poly-gamma-glutamate Tail of F420 Requires Both Domains of the F420: gamma-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
2BPQ
 
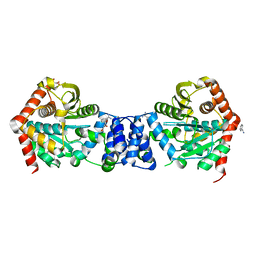 | | Anthranilate phosphoribosyltransferase (TrpD) from Mycobacterium tuberculosis (Apo structure) | | Descriptor: | ANTHRANILATE PHOSPHORIBOSYLTRANSFERASE, BENZAMIDINE, GLYCEROL | | Authors: | Lee, C.E, Goodfellow, C, Javid-Majd, F, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-04-22 | | Release date: | 2006-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Trpd, a Metabolic Enzyme Essential for Lung Colonization by Mycobacterium Tuberculosis, in Complex with its Substrate Phosphoribosylpyrophosphate
J.Mol.Biol., 355, 2006
|
|
2CEV
 
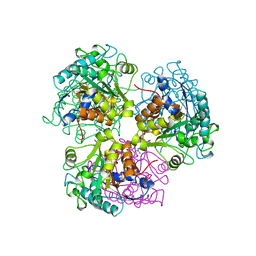 | | ARGINASE FROM BACILLUS CALDEVELOX, NATIVE STRUCTURE AT PH 8.5 | | Descriptor: | GUANIDINE, MANGANESE (II) ION, PROTEIN (ARGINASE) | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-10 | | Release date: | 1999-04-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
4A1O
 
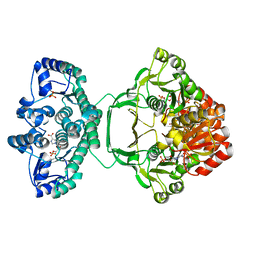 | | Crystal structure of Mycobacterium tuberculosis PurH complexed with AICAR and a novel nucleotide CFAIR, at 2.48 A resolution. | | Descriptor: | 5-(FORMYLAMINO)-1-(5-O-PHOSPHONO-BETA-D-RIBOFURANOSYL)-1H-IMIDAZOLE-4-CARBOXYLIC ACID, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, BIFUNCTIONAL PURINE BIOSYNTHESIS PROTEIN PURH, ... | | Authors: | Le Nours, J, Bulloch, E.M.M, Zhang, Z, Greenwood, D.R, Middleditch, M.J, Dickson, J.M.J, Baker, E.N. | | Deposit date: | 2011-09-17 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural Analyses of a Purine Biosynthetic Enzyme from Mycobacterium Tuberculosis Reveal a Novel Bound Nucleotide.
J.Biol.Chem., 286, 2011
|
|
4BC4
 
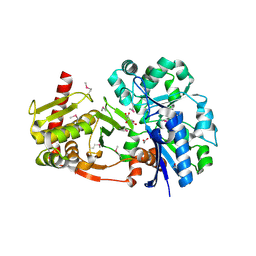 | | Crystal structure of human D-xylulokinase in complex with D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, D-XYLULOSE, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
4BC5
 
 | | Crystal structure of human D-xylulokinase in complex with inhibitor 5- deoxy-5-fluoro-D-xylulose | | Descriptor: | 1,2-ETHANEDIOL, 5-deoxy-5-fluoro-D-xylulose, XYLULOSE KINASE | | Authors: | Bunker, R.D, Loomes, K.M, Baker, E.N. | | Deposit date: | 2012-10-01 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Function of Human Xylulokinase, an Enzyme with Important Roles in Carbohydrate Metabolism
J.Biol.Chem., 288, 2013
|
|
