3UXA
 
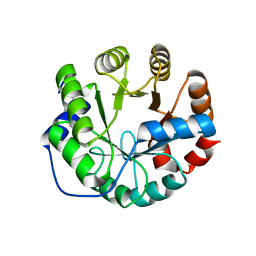 | | Designed protein KE59 R1 7/10H | | 分子名称: | Kemp eliminase KE59 R1 7/10H, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-05 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UZ5
 
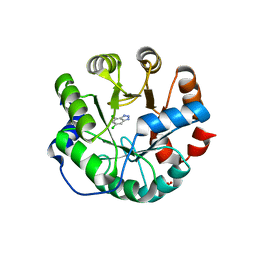 | | Designed protein KE59 R13 3/11H | | 分子名称: | 5,7-dichloro-1H-benzotriazole, Kemp eliminase KE59 R13 3/11H, PHOSPHATE ION, ... | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-07 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8GLT
 
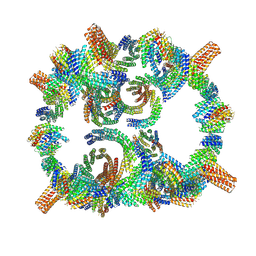 | | Backbone model of de novo-designed chlorophyll-binding nanocage O32-15 | | 分子名称: | C2-chlorophyll-comp_O32-15_ctermHis, polyalanine model, C3-comp_O32-15 | | 著者 | Redler, R.L, Ennist, N.M, Wang, S, Baker, D, Ekiert, D.C, Bhabha, G. | | 登録日 | 2023-03-23 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (6.5 Å) | | 主引用文献 | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 20, 2024
|
|
6TJ1
 
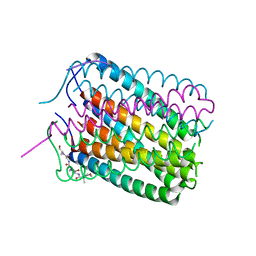 | | Crystal structure of a de novo designed hexameric helical-bundle protein | | 分子名称: | De novo designed WSHC6, purification tag | | 著者 | Xu, C, Pei, X.Y, Luisi, B.F, Baker, D. | | 登録日 | 2019-11-23 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Computational design of transmembrane pores.
Nature, 585, 2020
|
|
3JVE
 
 | |
4NWR
 
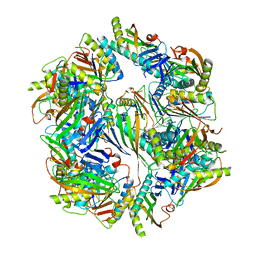 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T33-28 | | 分子名称: | Macrophage migration inhibitory factor-like protein, integron gene cassette protein | | 著者 | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2013-12-06 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
4NWO
 
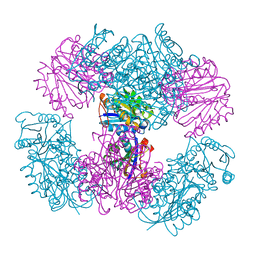 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage T33-15 | | 分子名称: | CALCIUM ION, Chorismate mutase AroH, Molybdenum cofactor biosynthesis protein MogA | | 著者 | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2013-12-06 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
3UYC
 
 | | Designed protein KE59 R8_2/7A | | 分子名称: | Kemp eliminase KE59 R8_2/7A, PHOSPHATE ION | | 著者 | Khersonsky, O, Kiss, G, Roethlisberger, D, Dym, O, Albeck, S, Houk, K.N, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-12-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Bridging the gaps in design methodologies by evolutionary optimization of the stability and proficiency of designed Kemp eliminase KE59.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4NWQ
 
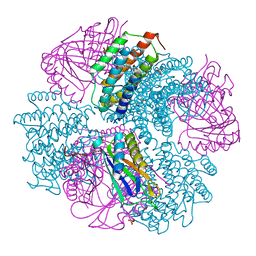 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group F4132 | | 分子名称: | Putative uncharacterized protein PH0671, SULFATE ION, Uncharacterized protein | | 著者 | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | 登録日 | 2013-12-06 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | 分子名称: | De Novo design cysteine esterase FR29 | | 著者 | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2011-09-30 | | 公開日 | 2011-12-07 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.797 Å) | | 主引用文献 | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3NPU
 
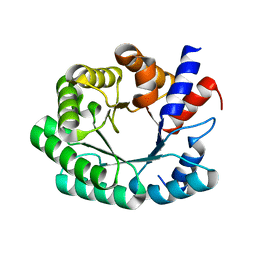 | | Optimization of the in silico designed Kemp eliminase KE70 by computational design and directed evolution | | 分子名称: | deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
3NPW
 
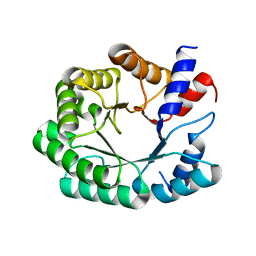 | | In silico designed of an improved Kemp eliminase KE70 mutant by computational design and directed evolution | | 分子名称: | deoxyribose phosphate aldolase | | 著者 | Khersonsky, O, Rothlisberge, D, Wollacott, A.M, Dym, O, Baker, D, Tawfik, D.S, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2010-06-29 | | 公開日 | 2011-02-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Optimization of the in-silico-designed kemp eliminase KE70 by computational design and directed evolution
J.Mol.Biol., 407, 2011
|
|
6BES
 
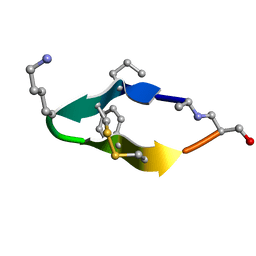 | | Solution structure of de novo macrocycle design11_ss | | 分子名称: | (DAL)Q(DPR)(DCY)(DLY)DS(DTY)(DCY)P(DSN) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-10-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6WRW
 
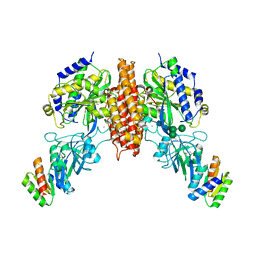 | | Crystal structure of computationally designed protein 2DS25.5 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 2DS25.5, ... | | 著者 | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRX
 
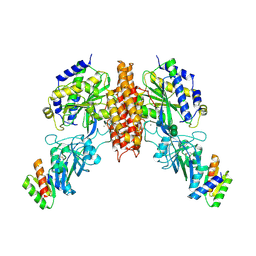 | | Crystal structure of computationally designed protein 2DS25.1 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Abraham, J, Coscia, A, Olal, D, Sahtoe, D.D, Baker, D, Clark, L. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6WRV
 
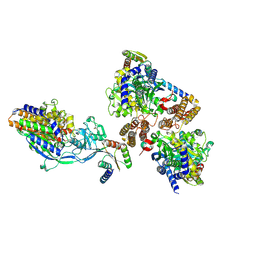 | | Crystal structure of computationally designed protein 3DS18 in complex with the human Transferrin receptor ectodomain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Computationally designed protein 3DS18, ... | | 著者 | Abraham, J, Baker, D, Sahtoe, D.D, Coscia, A, Clark, L, Olal, D. | | 登録日 | 2020-04-30 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Transferrin receptor targeting by de novo sheet extension.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8GAB
 
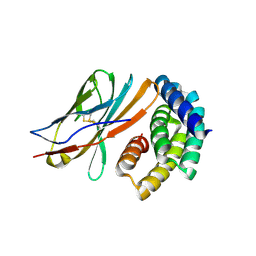 | | Crystal structure of CTLA-4 in complex with a high affinity CTLA-4 binder | | 分子名称: | CTLA-4 binder, Cytotoxic T-lymphocyte protein 4, POTASSIUM ION | | 著者 | Yang, W, Almo, S.C, Baker, D, Ghosh, A. | | 登録日 | 2023-02-22 | | 公開日 | 2024-08-21 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Design of High Affinity Binders to Convex Protein Target Sites.
Biorxiv, 2024
|
|
6BEW
 
 | | Solution structure of de novo macrocycle design7.2 | | 分子名称: | (DHI)P(DAS)(DGN)(DSN)(DGL)P | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BET
 
 | | Solution structure of de novo macrocycle design12_ss | | 分子名称: | H(DPR)(DVA)CIP(DPR)E(DLY)VC(DGL) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-10-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BER
 
 | | Solution structure of de novo macrocycle design10.2 | | 分子名称: | E(DVA)DP(DGL)(DHI)(DPR)N(DAL)(DPR) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEQ
 
 | | Solution structure of de novo macrocycle design10.1 | | 分子名称: | AAR(DVA)(DPR)R(DLE)(DTH)PE | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2018-01-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
6BEU
 
 | | Solution structure of de novo macrocycle design14_ss | | 分子名称: | (DCY)N(DVA)(DPR)DVYC(DPR)(DSG)KY(DVA)(DPR) | | 著者 | Shortridge, M.D, Hosseinzadeh, P, Pardo-Avila, F, Varani, G, Baker, D. | | 登録日 | 2017-10-25 | | 公開日 | 2017-12-27 | | 最終更新日 | 2024-10-30 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Comprehensive computational design of ordered peptide macrocycles.
Science, 358, 2017
|
|
2MDW
 
 | |
3Q9U
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | 分子名称: | COENZYME A, CoA binding protein, consensus ankyrin repeat | | 著者 | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | 登録日 | 2011-01-10 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
5HRY
 
 | | Computationally Designed Cyclic Dimer ank3C2_1 | | 分子名称: | ank3C2_1 | | 著者 | Cascio, D, McNamara, D.E, Fallas, J.A, Baker, D, Yeates, T.O. | | 登録日 | 2016-01-24 | | 公開日 | 2017-04-12 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
