4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2014-04-10 | | 公開日 | 2014-05-07 | | 最終更新日 | 2017-03-08 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
3NYI
 
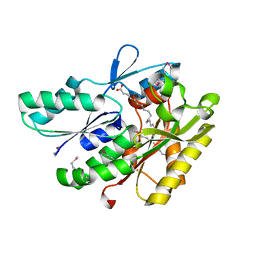 | | The crystal structure of a fat acid (stearic acid)-binding protein from Eubacterium ventriosum ATCC 27560. | | 分子名称: | STEARIC ACID, fat acid-binding protein | | 著者 | Zhang, R, Tan, K, Li, H, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-07-15 | | 公開日 | 2010-09-22 | | 最終更新日 | 2016-12-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The crystal structure of a fat acid (stearic acid)-binding protein from Eubacterium ventriosum ATCC 27560.
TO BE PUBLISHED
|
|
3M46
 
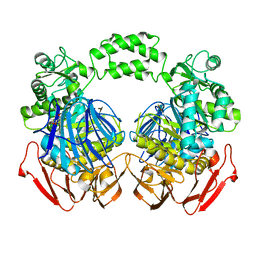 | | The crystal structure of the D73A mutant of glycoside HYDROLASE (FAMILY 31) from Ruminococcus obeum ATCC 29174 | | 分子名称: | GLYCEROL, Uncharacterized protein | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-10 | | 公開日 | 2010-03-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch
Faseb J., 24, 2010
|
|
7TKV
 
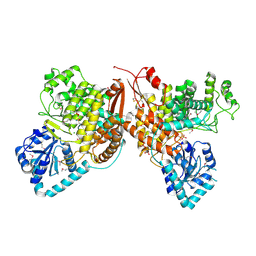 | | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus | | 分子名称: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Kim, Y, Crawford, M, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-17 | | 公開日 | 2022-01-26 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Thioredox_DsbH Domain-Containing Uncharacterized Protein Bab1_2064 from Brucella abortus
To Be Published
|
|
7TJ1
 
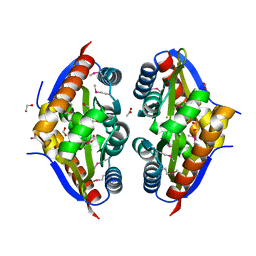 | | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Kim, Y, Tesar, C, Pastore, T, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2022-01-14 | | 公開日 | 2022-01-26 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Putative Fluoride Ion Transporter CrcB Bab1_1389 from Brucella abortus
To Be Published
|
|
3M6D
 
 | | The crystal structure of the d307a mutant of glycoside Hydrolase (family 31) from ruminococcus obeum atcc 29174 | | 分子名称: | Uncharacterized protein | | 著者 | Tan, K, Tesar, C, Freeman, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-03-15 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Novel alpha-glucosidase from human gut microbiome: substrate specificities and their switch.
Faseb J., 24, 2010
|
|
5C6U
 
 | | Rv3722c aminotransferase from Mycobacterium tuberculosis | | 分子名称: | Aminotransferase, CHLORIDE ION, PHOSPHATE ION, ... | | 著者 | OSIPIUK, J, Hatzos-Skintges, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2015-06-23 | | 公開日 | 2015-07-15 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Rv3722c aminotransferase from Mycobacterium tuberculosis.
to be published
|
|
7SF6
 
 | | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense | | 分子名称: | 1,2-ETHANEDIOL, 2-(2,3-DIHYDROXY-BENZOYLAMINO)-3-HYDROXY-PROPIONIC ACID, CHLORIDE ION, ... | | 著者 | Kim, Y, Patel, H.P, Nordquist, K.A, Schaab, K.M, Sha, J, Babnigg, G, Bond, A.H, Joachimiak, A, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2021-10-03 | | 公開日 | 2021-12-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Crystal Structure of Siderophore Binding Protein FatB from Desulfitobacterium hafniense
To Be Published
|
|
4OVM
 
 | | Crystal structure of SgcJ protein from Streptomyces carzinostaticus | | 分子名称: | uncharacterized protein SgcJ | | 著者 | Chang, C, Bigelow, L, Clancy, S, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2013-11-20 | | 公開日 | 2013-12-25 | | 最終更新日 | 2023-03-22 | | 実験手法 | X-RAY DIFFRACTION (2.719 Å) | | 主引用文献 | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J.Antibiot., 2016
|
|
4HX6
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 | | 分子名称: | ACETATE ION, Oxidoreductase, SULFATE ION | | 著者 | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-09 | | 公開日 | 2012-11-28 | | 最終更新日 | 2016-12-07 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
5UVE
 
 | | Crystal Structure of the ABC Transporter Substrate-binding protein BAB1_0226 from Brucella abortus | | 分子名称: | CALCIUM ION, GLYCEROL, Substrate-binding region of ABC-type glycine betaine transport system | | 著者 | Kim, Y, Chhor, G, Endres, M, Hero, J, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2017-02-20 | | 公開日 | 2017-03-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal Structure of Beta-barrel-like Protein of Unknown Function
To Be Published
|
|
6WRH
 
 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant | | 分子名称: | CHLORIDE ION, GLYCEROL, Non-structural protein 3, ... | | 著者 | Osipiuk, J, Tesar, C, Jedrzejczak, R, Endres, M, Welk, L, Babnigg, G, Kim, Y, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-29 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
5EVL
 
 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum | | 分子名称: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-11-19 | | 公開日 | 2015-12-02 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum
To Be Published
|
|
5EVI
 
 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae | | 分子名称: | 1,2-ETHANEDIOL, Beta-Lactamase/D-Alanine Carboxypeptidase, SULFATE ION | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-11-19 | | 公開日 | 2016-01-13 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae
To Be Published
|
|
4RAM
 
 | | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Penicillin G | | 分子名称: | Beta-lactamase NDM-1, CHLORIDE ION, OPEN FORM - PENICILLIN G, ... | | 著者 | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | 登録日 | 2014-09-10 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.495 Å) | | 主引用文献 | Crystal Structure of New Delhi Metallo-beta-Lactamase-1 Mutant M67V Complexed with Hydrolyzed Penicillin G
To be Published
|
|
5E2H
 
 | | Crystal Structure of D-alanine Carboxypeptidase AmpC from Mycobacterium smegmatis | | 分子名称: | Beta-lactamase, CHLORIDE ION, GLYCEROL | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-01 | | 公開日 | 2015-10-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of D-alanine Carboxypeptidase AmpC from Mycobacterium smegmatis
To Be Published
|
|
5E43
 
 | | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum | | 分子名称: | ACETATE ION, Beta-lactamase, NITRATE ION | | 著者 | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2015-10-05 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.7095 Å) | | 主引用文献 | Crystal Structure of Beta-lactamase Sros_5706 from Streptosporangium roseum
To Be Published
|
|
4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | 分子名称: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | 著者 | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-04-07 | | 公開日 | 2014-05-07 | | 最終更新日 | 2015-02-11 | | 実験手法 | X-RAY DIFFRACTION (1.349 Å) | | 主引用文献 | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | 分子名称: | SULFATE ION, TlmII | | 著者 | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-06 | | 公開日 | 2012-11-21 | | 最終更新日 | 2014-04-16 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | 著者 | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-12-06 | | 公開日 | 2013-02-20 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | 分子名称: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | 著者 | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2012-11-20 | | 公開日 | 2012-12-05 | | 最終更新日 | 2013-01-30 | | 実験手法 | X-RAY DIFFRACTION (2.148 Å) | | 主引用文献 | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
5UPQ
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP465 ligand | | 分子名称: | 5'-O-[(R)-{[(7beta,8alpha,9beta,10alpha,13alpha,16beta)-7,16-dihydroxy-18-oxokauran-18-yl]oxy}(hydroxy)phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-02-03 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.42 Å) | | 主引用文献 | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
5UPS
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP663 ligand | | 分子名称: | 5'-O-[(R)-hydroxy{[(7beta,8alpha,9beta,10alpha,11beta,13alpha)-7-hydroxy-19-oxo-11,16-epoxykauran-19-yl]oxy}phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, FORMIC ACID, ... | | 著者 | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.-Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-02-03 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
3UO3
 
 | | Jac1 co-chaperone from Saccharomyces cerevisiae, 5-182 clone | | 分子名称: | ACETATE ION, J-type co-chaperone JAC1, mitochondrial | | 著者 | Osipiuk, J, Bigelow, L, Mulligan, R, Feldmann, B, Babnigg, G, Marszalek, J, Craig, E.A, Dutkiewicz, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2011-11-16 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Interaction of j-protein co-chaperone jac1 with fe-s scaffold isu is indispensable in vivo and conserved in evolution.
J.Mol.Biol., 417, 2012
|
|
5UPT
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP468 ligand | | 分子名称: | (7alpha,8alpha,10alpha,13alpha)-7,16-dihydroxykauran-18-oic acid, Acyl-CoA synthetase PtmA2, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2017-02-03 | | 公開日 | 2017-02-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
