3NYI
 
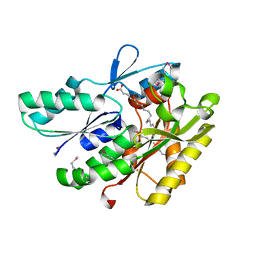 | | The crystal structure of a fat acid (stearic acid)-binding protein from Eubacterium ventriosum ATCC 27560. | | Descriptor: | STEARIC ACID, fat acid-binding protein | | Authors: | Zhang, R, Tan, K, Li, H, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a fat acid (stearic acid)-binding protein from Eubacterium ventriosum ATCC 27560.
TO BE PUBLISHED
|
|
6MGZ
 
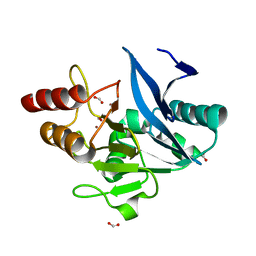 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae | | Descriptor: | FORMIC ACID, MAGNESIUM ION, NDM-4, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.647 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 4 from Klebsiella pneumoniae
To Be Published
|
|
6MGY
 
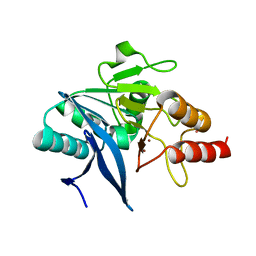 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae | | Descriptor: | GLYCEROL, Metallo-beta-lactamase NDM-5, POTASSIUM ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 5 from Klebsiella pneumoniae
To Be Published
|
|
6NHU
 
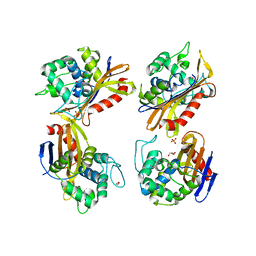 | | Crystal Structure of the Beta Lactamase Class D YbxI from Agrobacterium fabrum | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, GLYCEROL, ... | | Authors: | Kim, Y, Welk, L, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbxI from Agrobacterium fabrum
To Be Published
|
|
6MH0
 
 | | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 3 from Klebsiella pneumoniae | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase NDM-3, SULFATE ION, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the New Deli Metallo Beta Lactamase Variant 3 from Klebsiella pneumoniae
To Be Published
|
|
6NJC
 
 | | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus | | Descriptor: | ACETIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Kim, Y, Li, H, Biglow, L, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-03 | | Release date: | 2019-01-16 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Sialate O-acetylesterase from Bacteroides vulgatus
To Be Published
|
|
6NTR
 
 | | Crystal Structure of Beta-barrel-like Protein of Domain of Unknown Function DUF1849 from Brucella abortus | | Descriptor: | 1,2-ETHANEDIOL, ATP/GTP-binding site-containing protein A, GLYCEROL | | Authors: | Kim, Y, Bigelow, L, Endres, M, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-01-30 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | BrucellaPeriplasmic Protein EipB Is a Molecular Determinant of Cell Envelope Integrity and Virulence.
J.Bacteriol., 201, 2019
|
|
6NHS
 
 | | Crystal Structure of the Beta Lactamase Class D YbXI from Nostoc | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Kim, Y, Tesar, C, Endres, M, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-23 | | Release date: | 2019-01-16 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbXI from Nostoc
To Be Published
|
|
6NI0
 
 | | Crystal Structure of the Beta Lactamase Class D YbxI from Burkholderia thailandensis | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Kim, Y, Wu, R, Endres, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Beta Lactamase Class D YbxI from Burkholderia thailandensis
To Be Published
|
|
6OK0
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | CHLORIDE ION, IMIDAZOLE, Sel1 repeat protein, ... | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-12 | | Release date: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (2.174 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6OYF
 
 | | The structure of condensation and adenylation domains of teixobactin-producing nonribosomal peptide synthetase Txo1 serine module | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, ... | | Authors: | Tan, K, Zhou, M, Jedrzejczak, R, Babnigg, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-14 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of teixobactin-producing nonribosomal peptide synthetase condensation and adenylation domains.
Curr Res Struct Biol, 2, 2020
|
|
4LPQ
 
 | | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894 | | Descriptor: | CHLORIDE ION, ErfK/YbiS/YcfS/YnhG family protein | | Authors: | Nocek, B, Bigelow, L, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-07-16 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of the L,D-transpeptidase (residues 123-326) from Xylanimonas cellulosilytica DSM 15894
To be Published
|
|
5UVE
 
 | | Crystal Structure of the ABC Transporter Substrate-binding protein BAB1_0226 from Brucella abortus | | Descriptor: | CALCIUM ION, GLYCEROL, Substrate-binding region of ABC-type glycine betaine transport system | | Authors: | Kim, Y, Chhor, G, Endres, M, Hero, J, Babnigg, G, Crosson, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Beta-barrel-like Protein of Unknown Function
To Be Published
|
|
3RHF
 
 | | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Feldmann, B, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1
TO BE PUBLISHED
|
|
7RSK
 
 | | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus | | Descriptor: | Glycosyl hydrolase family 2, sugar binding domain protein | | Authors: | Kim, Y, Nocek, B, Endres, M, Joachimiak, G, Johnson, J, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2021-08-11 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure from microfluidic crystals of glycosyl hydrolase family 2 (GH2) member from Bacteroides cellulosilyticus
To Be Published
|
|
5UPS
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP663 ligand | | Descriptor: | 5'-O-[(R)-hydroxy{[(7beta,8alpha,9beta,10alpha,11beta,13alpha)-7-hydroxy-19-oxo-11,16-epoxykauran-19-yl]oxy}phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, FORMIC ACID, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.-Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
5UPQ
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP465 ligand | | Descriptor: | 5'-O-[(R)-{[(7beta,8alpha,9beta,10alpha,13alpha,16beta)-7,16-dihydroxy-18-oxokauran-18-yl]oxy}(hydroxy)phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
5V4F
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis | | Descriptor: | GLYCEROL, Putative translational inhibitor protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfB from Yersinia pestis
To Be Published
|
|
3PHA
 
 | | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-glucosidase | | Authors: | Tan, K, Tesar, C, Keigher, L, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-11-03 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | The crystal structure of the W169Y mutant of alpha-glucosidase (gh31 family) from Ruminococcus obeum atcc 29174 in complex with acarbose
To be Published
|
|
7TE5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis | | Descriptor: | MAGNESIUM ION, Pirin family protein Yhak | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-04 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK from Yersinia pestis
To Be Published
|
|
7TEM
 
 | | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of the Putative Exported Protein YPO2471 from Yersinia pestis
To Be Published
|
|
5V4D
 
 | | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis | | Descriptor: | ACETIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Kim, Y, Chhor, G, Endres, M, Krishnan, A, Babnigg, G, Schneewind, O, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-09 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Protein of Unknown Function of the Conserved Rid Protein Family YyfA from Yersinia pestis
To Be Published
|
|
3RKJ
 
 | | Crystal Structure of New Delhi Metallo-Beta-Lactamase-1 from Klebsiella pnueumoniae | | Descriptor: | Beta-lactamase NDM-1, GLYCEROL, SULFATE ION | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Binkowski, T.A, Babnigg, G, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
7THW
 
 | | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Putative OmpA-family membrane protein | | Authors: | Kim, Y, Tesar, C, Chhor, G, Clancy, S, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-12 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Soluble Domain of the Putative OmpA -Family Membrane Protein YPO0514 from Yersinia pestis
To Be Published
|
|
7TG5
 
 | | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the Presence of Fe Ion from Yersinia pestis | | Descriptor: | CHLORIDE ION, FE (III) ION, Pirin family protein | | Authors: | Kim, Y, Chhor, G, Endres, M, Babnigg, G, Schneewind, O, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-07 | | Release date: | 2022-01-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of the Pirin Family Protein Redox-sensitive Bicupin YhaK in the presence of Fe ion from Yersinia pestis
To Be Published
|
|
