2IZI
 
 | | STREPTAVIDIN-BIOTIN PH 2.53 I4122 STRUCTURE | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
2IZH
 
 | | STREPTAVIDIN-BIOTIN PH 10.44 I222 COMPLEX | | Descriptor: | BIOTIN, STREPTAVIDIN | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
3A0K
 
 | | Crystal structure of an antiflamatory legume lectin from Cymbosema roseum seeds | | Descriptor: | ALPHA-AMINOBUTYRIC ACID, CALCIUM ION, Cymbosema roseum mannose-specific lectin, ... | | Authors: | Rocha, B.A.M, Delatorre, P, Marinho, E.S, Benevides, R.G, Moura, T.R, Souza, L.A.G, Nascimento, K.S, Sampaio, A.H, Cavada, B.S. | | Deposit date: | 2009-03-20 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for both pro- and anti-inflammatory response induced by mannose-specific legume lectin from Cymbosema roseum
Biochimie, 2011
|
|
4LND
 
 | | Crystal structure of human apurinic/apyrimidinic endonuclease 1 with essential Mg2+ cofactor | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Manvilla, B.A, Pozharski, E, Toth, E.A, Drohat, A.C. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg(2+) cofactor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1K1R
 
 | | HETERODUPLEX OF CHIRALLY PURE R-METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(RMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenkova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
1K1H
 
 | | HETERODUPLEX OF CHIRALLY PURE METHYLPHOSPHONATE/DNA DUPLEX | | Descriptor: | 5'-D(*CP*(CMR)P*(RMP)P*(RMP)P*(SMP)P*(CMR)P*(RMP))-3', 5'-D(*TP*GP*TP*TP*TP*GP*GP*C)-3' | | Authors: | Thiviyanathan, V, Vyazovkina, K.V, Gozansky, E.K, Bichenchova, E, Abramova, T.V, Luxon, B.A, Lebedev, A.V, Gorenstein, D.G. | | Deposit date: | 2001-09-25 | | Release date: | 2002-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry.
Biochemistry, 41, 2002
|
|
2IZG
 
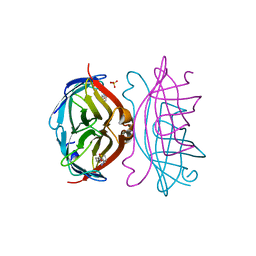 | | STREPTAVIDIN-BIOTIN PH 2.0 I222 COMPLEX | | Descriptor: | BIOTIN, STREPTAVIDIN, SULFATE ION | | Authors: | Katz, B.A. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Binding of biotin to streptavidin stabilizes intersubunit salt bridges between Asp61 and His87 at low pH.
J.Mol.Biol., 274, 1997
|
|
4Q2P
 
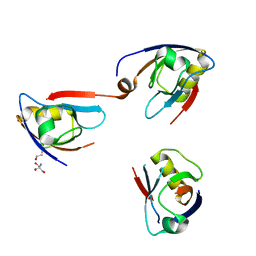 | | NHERF3 PDZ2 in Complex with a Phage-Derived Peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | | Authors: | Appleton, B.A, Wiesmann, C. | | Deposit date: | 2014-04-09 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural portrait of the PDZ domain family.
J.Mol.Biol., 426, 2014
|
|
4Q2N
 
 | |
4QX0
 
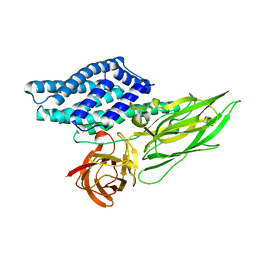 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2LKX
 
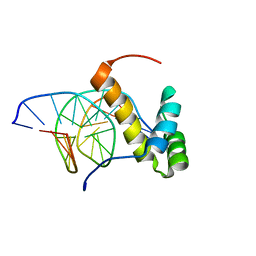 | | NMR structure of the homeodomain of Pitx2 in complex with a TAATCC DNA binding site | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*AP*TP*TP*AP*GP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*CP*TP*AP*AP*TP*CP*CP*CP*CP*G)-3'), Pituitary homeobox 3 | | Authors: | Baird-Titus, J.M, Doerdelmann, T, Chaney, B.A, Clark-Baldwin, K, Dave, V, Ma, J. | | Deposit date: | 2011-10-21 | | Release date: | 2012-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the K50 class homeodomain PITX2 bound to DNA and implications for mutations that cause Rieger syndrome
Biochemistry, 44, 2005
|
|
1MN3
 
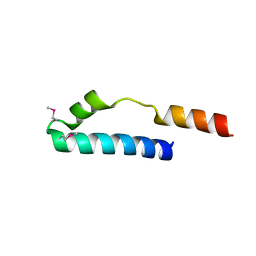 | | Cue domain of yeast Vps9p | | Descriptor: | Vacuolar protein sorting-associated protein VPS9 | | Authors: | Prag, G, Misra, S, Jones, E, Ghirlando, R, Davies, B.A, Horazdovsky, B.F, Hurley, J.H. | | Deposit date: | 2002-09-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanism of Ubiquitin Recognition by the CUE Domain of Vps9p
Cell(Cambridge,Mass.), 113, 2003
|
|
4F52
 
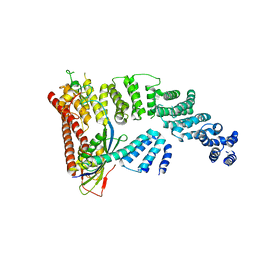 | | Structure of a Glomulin-RBX1-CUL1 complex | | Descriptor: | Cullin-1, E3 ubiquitin-protein ligase RBX1, Glomulin, ... | | Authors: | Duda, D.M, Olszewski, J.L, Schulman, B.A. | | Deposit date: | 2012-05-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a Glomulin-RBX1-CUL1 Complex: Inhibition of a RING E3 Ligase through Masking of Its E2-Binding Surface.
Mol.Cell, 47, 2012
|
|
4F4L
 
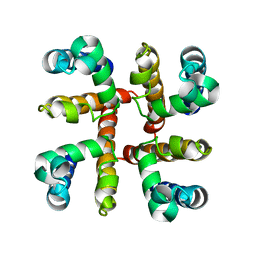 | | Open Channel Conformation of a Voltage Gated Sodium Channel | | Descriptor: | Ion transport protein | | Authors: | McCusker, E.C, Bagneris, C, Naylor, C.E, Cole, A.R, D'Avanzo, N, Nichols, C.G, Wallace, B.A. | | Deposit date: | 2012-05-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing.
Nat Commun, 3, 2012
|
|
4R2Y
 
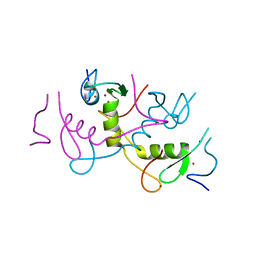 | | Crystal structure of APC11 RING domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissmann, F, Jarvis, M.A, Vanderlinden, R, Grace, C.R.R, Frye, J.J, Dube, P, Qiao, R, Petzold, G, Cho, S.E, Alsharif, O, Bao, J, Zheng, J, Nourse, A, Kurinov, I, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
4F2D
 
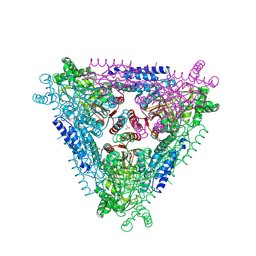 | | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol | | Descriptor: | ACETIC ACID, D-ribitol, L-arabinose isomerase, ... | | Authors: | Manjasetty, B.A, Burley, S.K, Almo, S.C, Chance, M.R, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-05-07 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli L-arabinose Isomerase (ECAI) complexed with Ribitol
TO BE PUBLISHED
|
|
1MHU
 
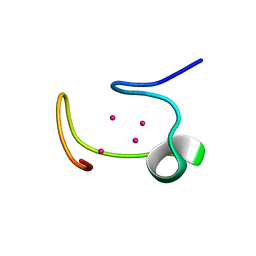 | | THE THREE-DIMENSIONAL STRUCTURE OF HUMAN [113CD7] METALLOTHIONEIN-2 IN SOLUTION DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | CADMIUM ION, CD7 METALLOTHIONEIN-2 | | Authors: | Braun, W, Messerle, B.A, Schaeffer, A, Vasak, M, Kaegi, J.H.R, Wuthrich, K. | | Deposit date: | 1990-05-14 | | Release date: | 1991-04-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of human [113Cd7]metallothionein-2 in solution determined by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
4RG7
 
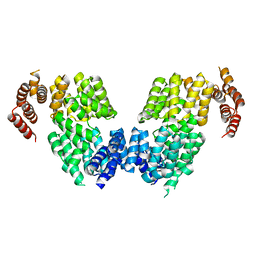 | | Crystal structure of APC3 | | Descriptor: | Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4F6N
 
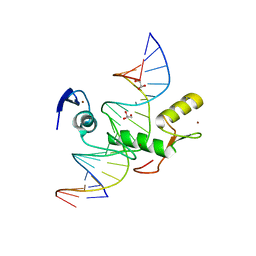 | | Crystal structure of Kaiso zinc finger DNA binding protein in complex with methylated CpG site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*TP*AP*GP*AP*(5CM)P*GP*(5CM)P*GP*GP*TP*GP*AP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*TP*CP*AP*CP*(5CM)P*GP*(5CM)P*GP*TP*CP*TP*AP*TP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4QX3
 
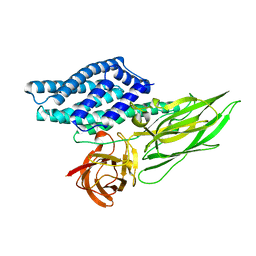 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1M24
 
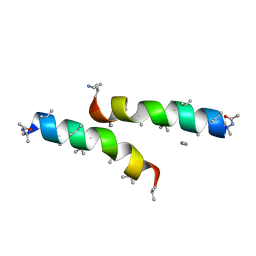 | |
1MIC
 
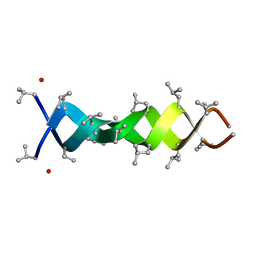 | |
4RF1
 
 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
4GAO
 
 | | DCNL complex with N-terminally acetylated NEDD8 E2 peptide | | Descriptor: | BROMIDE ION, DCN1-like protein 2, NEDD8-conjugating enzyme Ubc12 | | Authors: | Monda, J.K, Scott, D.C, Miller, D.J, Harper, J.W, Bennett, E.J, Schulman, B.A. | | Deposit date: | 2012-07-25 | | Release date: | 2012-11-28 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Conservation of Distinctive N-terminal Acetylation-Dependent Interactions across a Family of Mammalian NEDD8 Ligation Enzymes.
Structure, 21, 2013
|
|
4QX2
 
 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
