2VWK
 
 | | Uracil Recognition in Archaeal DNA Polymerases Captured by X-ray Crystallography. V93Q polymerase variant | | Descriptor: | DNA POLYMERASE, SODIUM ION, SULFATE ION | | Authors: | Firbank, S.J, Wardle, J, Heslop, P, Lewis, R.J, Connolly, B.A. | | Deposit date: | 2008-06-26 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Uracil Recognition in Archaeal DNA Polymerases Captured by X-Ray Crystallography.
J.Mol.Biol., 381, 2008
|
|
2WAB
 
 | | Structure of an active site mutant of a family two carbohydrate esterase from Clostridium thermocellum in complex with celluohexase | | Descriptor: | ENDOGLUCANASE E, GLYCEROL, IODIDE ION, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
1RMU
 
 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
1GJB
 
 | | ENGINEERING INHIBITORS HIGHLY SELECTIVE FOR THE S1 SITES OF SER190 TRYPSIN-LIKE SERINE PROTEASE DRUG TARGETS | | Descriptor: | 2-(2-HYDROXY-BIPHENYL)-1H-BENZOIMIDAZOLE-5-CARBOXAMIDINE, CITRIC ACID, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Katz, B.A, Sprengeler, P.A, Luong, C, Verner, E, Spencer, J.R, Breitenbucher, J.G, Hui, H, McGee, D, Allen, D, Martelli, A, Mackman, R.L. | | Deposit date: | 2001-04-27 | | Release date: | 2002-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets.
Chem.Biol., 8, 2001
|
|
6OP6
 
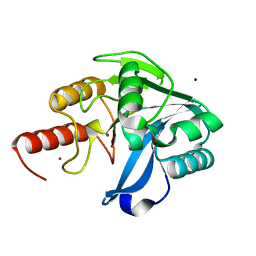 | | Structure of VIM-20 in the reduced state | | Descriptor: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
8QYF
 
 | |
8QYO
 
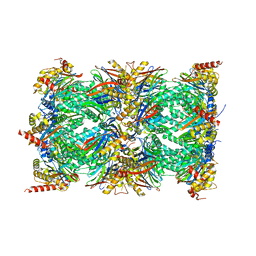 | | Human proteasome 20S core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYL
 
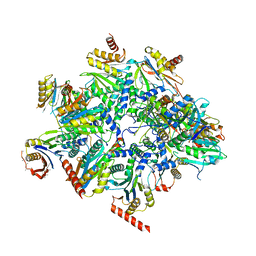 | | Human 20S proteasome assembly intermediate structure 2 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYM
 
 | | Human 20S proteasome assembly intermediate structure 3 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QYN
 
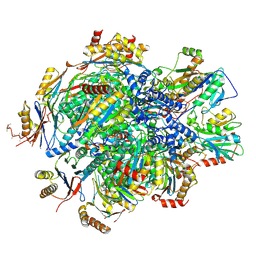 | | Human 20S proteasome assembly intermediate structure 5 | | Descriptor: | Proteasome assembly chaperone 1, Proteasome assembly chaperone 2, Proteasome maturation protein, ... | | Authors: | Schulman, B.A, Hanna, J.W, Harper, J.W, Adolf, F, Du, J, Rawson, S.D, Walsh Jr, R.M, Goodall, E.A. | | Deposit date: | 2023-10-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Visualizing chaperone-mediated multistep assembly of the human 20S proteasome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6ORM
 
 | | Crystal Structure of Peruvianin-I (Cysteine peptidase from Thevetia peruviana latex) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peruvianin-I, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cruz, W.T, Bezerra, E.H.S, da Silva, F.M.S, Freire, V.N, Ramos, M.V, Rocha, B.A.M, Freitas, C.D.T. | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure and specific location of a germin-like protein with proteolytic activity from Thevetia peruviana.
Plant Sci., 298, 2020
|
|
8Q7E
 
 | |
8Q3L
 
 | |
8Q7H
 
 | | Structure of CUL9-RBX1 ubiquitin E3 ligase complex in unneddylated and neddylated conformation - focused cullin dimer | | Descriptor: | Cullin-9, E3 ubiquitin-protein ligase RBX1, NEDD8, ... | | Authors: | Hopf, L.V.M, Horn-Ghetko, D, Schulman, B.A. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Noncanonical assembly, neddylation and chimeric cullin-RING/RBR ubiquitylation by the 1.8 MDa CUL9 E3 ligase complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5N16
 
 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 1) | | Descriptor: | 5-cyclopropyl-2-(5-pyrazin-2-yl-1,2,4-oxadiazol-3-yl)benzo[b][1,4]benzothiazepin-6-one, Bromodomain-containing factor 1, GLYCEROL, ... | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
1HEN
 
 | |
1HEP
 
 | |
8QBN
 
 | |
6OP7
 
 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
8QE8
 
 | | Structure of the non-canonical CTLH E3 substrate receptor WDR26 bound to NMNAT1 substrate | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, Nicotinamide/nicotinic acid mononucleotide adenylyltransferase 1, WD repeat-containing protein 26, ... | | Authors: | Chrustowicz, J, Sherpa, D, Schulman, B.A. | | Deposit date: | 2023-08-30 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical substrate recognition by the human WDR26-CTLH E3 ligase regulates prodrug metabolism.
Mol.Cell, 84, 2024
|
|
6P5W
 
 | | Structure of DCN1 bound to 3-methyl-N-((4S,5S)-3-methyl-6-oxo-1-phenyl-4-(p-tolyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)benzamide | | Descriptor: | 3-methyl-N-[(4S,5S)-3-methyl-4-(4-methylphenyl)-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]benzamide, Lysozyme,DCN1-like protein 1 chimera | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
7LMW
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 7-methyl-3-(1~{H}-pyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-06 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
7LML
 
 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-(((3-(4-Carboxyphenoxy)benzyl)oxy)methyl)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 6-iodanyl-1~{H}-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2021-02-05 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
5N13
 
 | | Second Bromodomain (BD2) from Candida albicans Bdf1 in the unbound form | | Descriptor: | Bromodomain-containing factor 1, GLYCEROL | | Authors: | Mietton, F, Ferri, E, Champlebouxm, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
5N17
 
 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 3) | | Descriptor: | (2~{S})-~{N}-(5-methyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)oxolane-2-carboxamide, Bromodomain-containing factor 1, SULFATE ION | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvon, M, d'Enfer, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
