5EYY
 
 | | Tetragonal Form of Centrolobium tomentosum seed lectin (CTL) complexed with Man1-3Man-OMe. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Centrolobium tomentosum lectin, ... | | Authors: | Pinto-Junior, V.R, Osterne, V.J.S, Santiago, M.Q, Almeida, A.C, Lossio, C.F, Silva-Filho, J.C, Almeida, R.P.H, Teixeira, C.S, Delatorre, P, Rocha, B.A.M, Nascimento, K.S, Cavada, B.S. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural analysis of Centrolobium tomentosum seed lectin with inflammatory activity.
Arch.Biochem.Biophys., 596, 2016
|
|
6CKW
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with (R)-3-((7-(((S)-2-amino-2-(2-methoxyphenyl)ethyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)-3-(3-chlorophenyl)propanenitrile | | Descriptor: | (3R)-3-[(7-{[(2S)-2-amino-2-(2-methoxyphenyl)ethyl]amino}-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]-3-(3-chlorophenyl)propanenitrile, DIMETHYL SULFOXIDE, Phosphopantetheine adenylyltransferase, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-03-01 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
6CHO
 
 | | Phosphopantetheine adenylyltransferase (CoaD) in complex with (R)-2-((1-(3-(4-methoxyphenoxy)phenyl)ethyl)amino)-5-methyl-[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one | | Descriptor: | 2-({(1R)-1-[3-(4-methoxyphenoxy)phenyl]ethyl}amino)-5-methyl[1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, Phosphopantetheine adenylyltransferase, SULFATE ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-22 | | Release date: | 2018-04-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Optimization of Phosphopantetheine Adenylyltransferase Inhibitors with Gram-Negative Antibacterial Activity.
J. Med. Chem., 61, 2018
|
|
6CCO
 
 | |
3BDY
 
 | | Dual specific bH1 Fab in complex with VEGF | | Descriptor: | Fab Fragment -Heavy Chain, Fab Fragment -Light Chain, GLYCEROL, ... | | Authors: | Bostrom, J.M, Wiesmann, C, Appleton, B.A. | | Deposit date: | 2007-11-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site
Science, 323, 2009
|
|
3RRD
 
 | | Native structure of Dioclea virgata lectin | | Descriptor: | CALCIUM ION, Lectin alpha chain, MANGANESE (II) ION | | Authors: | Delatorre, P, Nobrega, R.B, Gadelha, C.A.A, Santi-Gadelha, T, Farias, D.L, Rocha, B.A.M, Cavada, B.S, Nagano, C.S, Bezerra, E.H.S, Bezerra, M.J, Alencar, K.L. | | Deposit date: | 2011-04-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of Dioclea virgata lectin: Relations between carbohydrate binding site and nitric oxide production.
Biochimie, 94, 2012
|
|
4YAE
 
 | | Crystal structure of LigL-apo form from Sphingobium sp. strain SYK-6 | | Descriptor: | C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
4YAI
 
 | | Crystal structure of LigL in complex with NADH and GGE from Sphingobium sp. strain SYK-6 | | Descriptor: | (1S,2R)-1-(4-hydroxy-3-methoxyphenyl)-2-(2-methoxyphenoxy)propane-1,3-diol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase, ... | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
1GLO
 
 | | Crystal Structure of Cys25Ser mutant of human cathepsin S | | Descriptor: | CATHEPSIN S | | Authors: | Turkenburg, J.P, Lamers, M.B.A.C, Brzozowski, A.M, Wright, L.M, Hubbard, R.E, Sturt, S.L, Williams, D.H. | | Deposit date: | 2001-08-31 | | Release date: | 2002-08-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a Cys25->Ser Mutant of Human Cathepsin Cathepsin S
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4YAC
 
 | | Crystal structure of LigO in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
4YAG
 
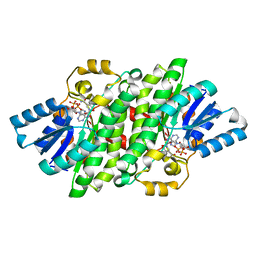 | | Crystal structure of LigL in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
4YAN
 
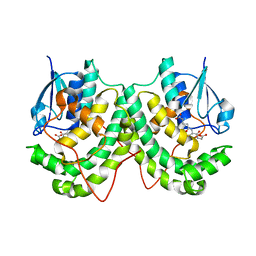 | | Crystal structure of LigE in complex with glutathione (GSH) from Sphingobium sp. strain SYK-6 | | Descriptor: | Beta-etherase, GLUTATHIONE | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of beta-Aryl Ether Bonds in Lignin.
J.Biol.Chem., 291, 2016
|
|
5D9K
 
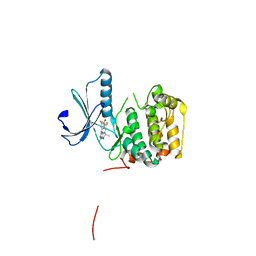 | | Rsk2 N-terminal Kinase in Complex with BI-D1870 | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-5,7-dimethyl-8-(3-methylbutyl)-7,8-dihydropteridin-6(5H)-one, GLYCEROL, Ribosomal protein S6 kinase alpha-3 | | Authors: | Appleton, B.A. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Potent and Selective RSK Inhibitors as Biological Probes.
J.Med.Chem., 58, 2015
|
|
5DG5
 
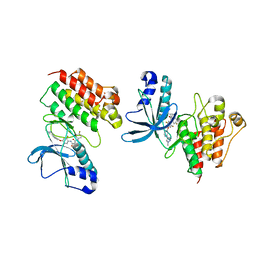 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET IN COMPLEX WITH ALTIRATINIB ANALOG DP-4157 | | Descriptor: | Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxam ide | | Authors: | Smith, B.D, Kaufman, M.D, Leary, C.B, Turner, B.A, Wise, S.A, Ahn, Y.M, Booth, R.J, Caldwell, T.M, Ensinger, C.L, Hood, M.M, Lu, W.-P, Patt, T.W, Patt, W.C, Rutkoski, T.J, Samarakoon, T, Telikepalli, H, Vogeti, L, Vogeti, S, Yates, K.M, Chun, L, Stewart, L.J, Clare, M, Flynn, D.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2.
Mol.Cancer Ther., 14, 2015
|
|
3B9V
 
 | |
3QTI
 
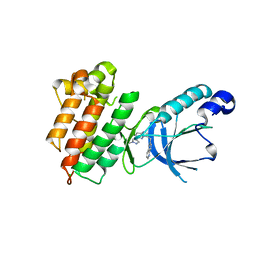 | | c-Met Kinase in Complex with NVP-BVU972 | | Descriptor: | 6-{[6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-b]pyridazin-3-yl]methyl}quinoline, CHLORIDE ION, GLYCEROL, ... | | Authors: | Appleton, B.A. | | Deposit date: | 2011-02-22 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Drug Resistance Screen Using a Selective MET Inhibitor Reveals a Spectrum of Mutations That Partially Overlap with Activating Mutations Found in Cancer Patients.
Cancer Res., 71, 2011
|
|
3EA2
 
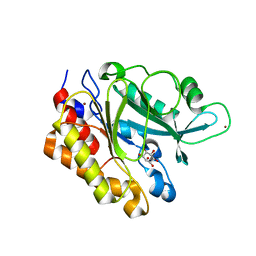 | | Crystal Structure of the Myo-inositol bound Y247S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, ZINC ION | | Authors: | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | Deposit date: | 2008-08-24 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Modulation of bacillus thuringiensis phosphatidylinositol-specific phospholipase C activity by mutations in the putative dimerization interface.
J.Biol.Chem., 284, 2009
|
|
3O2P
 
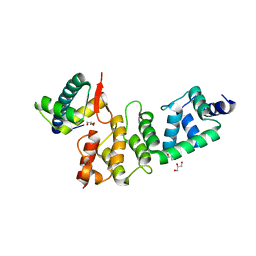 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1, GLYCEROL | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.233 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
5EYX
 
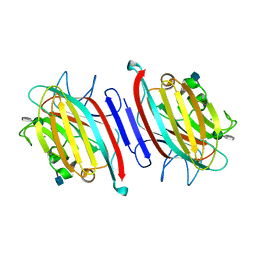 | | Monoclinic Form of Centrolobium tomentosum seed lectin (CTL) complexed with Man1-3Man-OMe. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Centrolobium tomentosum lectin, ... | | Authors: | Almeida, A.C, Osterne, V.J.S, Santiago, M.Q, Pinto-Junior, V.R, Silva-Filho, J.C, Lossio, C.F, Almeida, R.P.H, Teixeira, C.S, Delatorre, P, Rocha, B.A.M, Santiago, K.S, Cavada, B.S. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis of Centrolobium tomentosum seed lectin with inflammatory activity.
Arch.Biochem.Biophys., 596, 2016
|
|
4Y9D
 
 | | Crystal structure of LigD in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
6E93
 
 | | Crystal Structure of ZBTB38 C-terminal Zinc Fingers 6-9 in complex with methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*TP*CP*AP*TP*(DCM)P*GP*GP*(DCM)P*GP*CP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*(DCM)P*GP*CP*(DCM)P*GP*AP*TP*GP*AP*GP*TP*GP*C)-3'), ZINC ION, ... | | Authors: | Hudson, N.O, Whitby, F.G, Buck-Koehntop, B.A. | | Deposit date: | 2018-07-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.747 Å) | | Cite: | Structural insights into methylated DNA recognition by the C-terminal zinc fingers of the DNA reader protein ZBTB38.
J. Biol. Chem., 293, 2018
|
|
4YAM
 
 | | Crystal structure of LigE-apo form from Sphingobium sp. strain SYK-6 | | Descriptor: | Beta-etherase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of beta-Aryl Ether Bonds in Lignin.
J.Biol.Chem., 291, 2016
|
|
5FDL
 
 | | Crystal Structure of K103N/Y181C Mutant HIV-1 Reverse Transcriptase (RT) in Complex with IDX899 | | Descriptor: | P51 Reverse transcriptase, P66 Reverse transcriptase, methyl (R)-(2-carbamoyl-5-chloro-1H-indol-3-yl)[3-(2-cyanoethyl)-5-methylphenyl]phosphinate | | Authors: | Dousson, C.B, Alexandre, F.-R, Convard, T, Fisher, M, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2015-12-16 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of the Aryl-phospho-indole IDX899, a Highly Potent Anti-HIV Non-nucleoside Reverse Transcriptase Inhibitor.
J.Med.Chem., 59, 2016
|
|
5FFR
 
 | | Crystal Structure of Surfactant Protein-A complexed with phosphocholine | | Descriptor: | CALCIUM ION, PHOSPHOCHOLINE, Pulmonary surfactant-associated protein A, ... | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
5FFT
 
 | | Crystal Structure of Surfactant Protein-A Y221A Mutant | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein A | | Authors: | Goh, B.C, Wu, H, Rynkiewicz, M.J, Schulten, K, Seaton, B.A, McCormack, F.X. | | Deposit date: | 2015-12-18 | | Release date: | 2016-07-06 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Elucidation of Lipid Binding Sites on Lung Surfactant Protein A Using X-ray Crystallography, Mutagenesis, and Molecular Dynamics Simulations.
Biochemistry, 55, 2016
|
|
