4Y98
 
 | | Crystal structure of LigD-apo form from Sphingobium sp. strain SYK-6 | | Descriptor: | C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
4YA6
 
 | | Crystal structure of LigO-apo form from Sphingobium sp. strain SYK-6 | | Descriptor: | C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
3TFS
 
 | | Ternary complex structure of DNA polymerase beta with a gapped DNA substrate and a, b dAMP(CFH)PP in the active site: Stereoselective binding of (S) isomer | | Descriptor: | 2'-deoxy-5'-O-[(S)-{(S)-fluoro[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}(hydroxy)phosphoryl]adenosine, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*TP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Chamberlain, B.T, Batra, V.K, Beard, W.A, Kadina, A.P, Shock, D.D, Kashemirov, B.A, McKenna, C.E, Goodman, M.F, Wilson, S.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Stereospecific Formation of a Ternary Complex of (S)-alpha, beta-Fluoromethylene-dATP with DNA Pol beta.
Chembiochem, 13, 2012
|
|
4Y9D
 
 | | Crystal structure of LigD in complex with NADH from Sphingobium sp. strain SYK-6 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, C alpha-dehydrogenase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Structural and Biochemical Characterization of the Early and Late Enzymes in the Lignin beta-Aryl Ether Cleavage Pathway from Sphingobium sp. SYK-6.
J.Biol.Chem., 291, 2016
|
|
3U2D
 
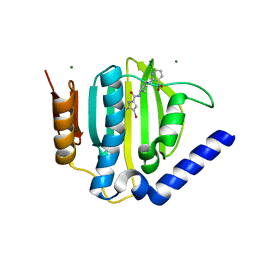 | | S. aureus GyrB ATPase domain in complex with small molecule inhibitor | | Descriptor: | 4-bromo-5-methyl-N-[1-(3-nitropyridin-2-yl)piperidin-4-yl]-1H-pyrrole-2-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
8Q7E
 
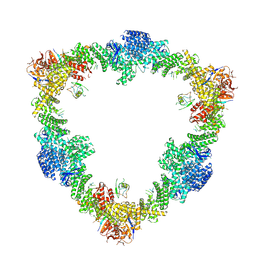 | |
8Q3L
 
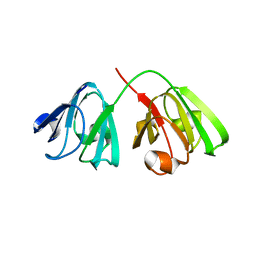 | |
4YAM
 
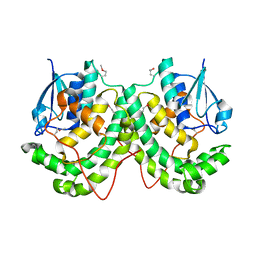 | | Crystal structure of LigE-apo form from Sphingobium sp. strain SYK-6 | | Descriptor: | Beta-etherase | | Authors: | Pereira, J.H, McAndrew, R.P, Heins, R.A, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2015-02-17 | | Release date: | 2015-12-16 | | Last modified: | 2016-07-20 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structural Basis of Stereospecificity in the Bacterial Enzymatic Cleavage of beta-Aryl Ether Bonds in Lignin.
J.Biol.Chem., 291, 2016
|
|
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | Descriptor: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | Deposit date: | 2023-07-11 | | Release date: | 2024-02-14 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3U2K
 
 | | S. aureus GyrB ATPase domain in complex with a small molecule inhibitor | | Descriptor: | 2-chloro-6-(4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}piperidin-1-yl)pyridine-4-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
3TVI
 
 | | Crystal structure of Clostridium acetobutylicum aspartate kinase (CaAK): An important allosteric enzyme for industrial amino acids production | | Descriptor: | ASPARTIC ACID, Aspartokinase, LYSINE | | Authors: | Manjasetty, B.A, Chance, M.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-09-20 | | Release date: | 2011-11-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Clostridium acetobutylicum Aspartate kinase (CaAK): An important allosteric enzyme for amino acids production.
Biotechnol Rep (Amst), 3, 2014
|
|
8UCQ
 
 | |
3O6B
 
 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) low resolution | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3T7E
 
 | | Atg8 transfer from Atg7 to Atg3: a distinctive E1-E2 architecture and mechanism in the autophagy pathway | | Descriptor: | Ubiquitin-like modifier-activating enzyme ATG7, ZINC ION | | Authors: | Taherbhoy, A.M, Tait, S.W, Kaiser, S.E, Williams, A.H, Deng, A, Nourse, A, Hammel, M, Kurinov, I, Rock, C.O, Green, D.R, Schulman, B.A. | | Deposit date: | 2011-07-30 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Atg8 transfer from atg7 to atg3: a distinctive e1-e2 architecture and mechanism in the autophagy pathway.
Mol.Cell, 44, 2011
|
|
3OKU
 
 | | Human Carbonic Anhydrase II in complex with 2-Ethylestrone-3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3OIM
 
 | | Human Carbonic anhydrase II bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3PBF
 
 | | Surfactant Protein-A neck and carbohydrate recognition domain (NCRD) complexed with glycerol | | Descriptor: | CALCIUM ION, GLYCEROL, Pulmonary surfactant-associated protein A | | Authors: | Shang, F, Rynkiewicz, M.J, McCormack, F.X, Wu, H, Cafarella, T.M, Head, J, Seaton, B.A. | | Deposit date: | 2010-10-20 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic complexes of surfactant protein A and carbohydrates reveal ligand-induced conformational change.
J.Biol.Chem., 286, 2011
|
|
3PFJ
 
 | | Crystal structure of Cel7A from Talaromyces emersonii | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiohydrolase 1 catalytic domain, SULFATE ION, ... | | Authors: | Qin, L, Pereira, J.H, McAndrew, R.P, Simmons, B.A, Sapra, R, Adams, P.D, Sale, K.L. | | Deposit date: | 2010-10-28 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3611 Å) | | Cite: |
|
|
3URC
 
 | | T181G mutant of alpha-Lytic Protease | | Descriptor: | Alpha-lytic protease, GLYCEROL, SULFATE ION | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2011-11-22 | | Release date: | 2012-05-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Functional modulation of a protein folding landscape via side-chain distortion.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RRD
 
 | | Native structure of Dioclea virgata lectin | | Descriptor: | CALCIUM ION, Lectin alpha chain, MANGANESE (II) ION | | Authors: | Delatorre, P, Nobrega, R.B, Gadelha, C.A.A, Santi-Gadelha, T, Farias, D.L, Rocha, B.A.M, Cavada, B.S, Nagano, C.S, Bezerra, E.H.S, Bezerra, M.J, Alencar, K.L. | | Deposit date: | 2011-04-29 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure of Dioclea virgata lectin: Relations between carbohydrate binding site and nitric oxide production.
Biochimie, 94, 2012
|
|
3U5Z
 
 | | Structure of T4 Bacteriophage clamp loader bound to the T4 clamp, primer-template DNA, and ATP analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
3U4X
 
 | | Crystal structure of a lectin from Camptosema pedicellatum seeds in complex with 5-bromo-4-chloro-3-indolyl-alpha-D-mannose | | Descriptor: | 5-bromo-4-chloro-1H-indol-3-yl alpha-D-mannopyranoside, CALCIUM ION, Camptosema pedicellatum lectin (CPL), ... | | Authors: | Rocha, B.A.M, Teixeira, C.S, Moura, T.R, Silva, H.C, Pereira-Junior, F.N, Nagano, C.S, Delatorre, P, Cavada, B.S. | | Deposit date: | 2011-10-10 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the lectin of Camptosema pedicellatum: implications of a conservative substitution at the hydrophobic subsite.
J.Biochem., 152, 2012
|
|
3TTZ
 
 | | Crystal structure of a topoisomerase ATPase inhibitor | | Descriptor: | 2-[(3S,4R)-4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}-3-fluoropiperidin-1-yl]-1,3-thiazole-5-carboxylic acid, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Read, J, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-09-15 | | Release date: | 2011-11-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: Optimization of antibacterial activity and efficacy.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3U60
 
 | | Structure of T4 Bacteriophage Clamp Loader Bound To Open Clamp, DNA and ATP Analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA polymerase accessory protein 44, DNA polymerase accessory protein 62, ... | | Authors: | Kelch, B.A, Makino, D.L, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | How a DNA polymerase clamp loader opens a sliding clamp.
Science, 334, 2011
|
|
3OIL
 
 | | Human Carbonic anhydrase II mutant A65S, N67Q (CA IX mimic) bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, ZINC ION | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
