7LSM
 
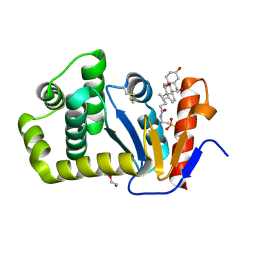 | | Crystal structure of E.coli DsbA in complex with bile salt taurocholate | | Descriptor: | DI(HYDROXYETHYL)ETHER, TAUROCHOLIC ACID, Thiol:disulfide interchange protein DsbA | | Authors: | Wang, G, Heras, B. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Selective Binding of Small Molecules to Vibrio cholerae DsbA Offers a Starting Point for the Design of Novel Antibacterials.
Chemmedchem, 17, 2022
|
|
6KYZ
 
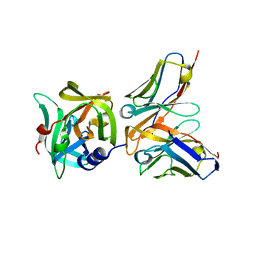 | | HRV14 3C in complex with single chain antibody YDF | | Descriptor: | Genome polyprotein, YDF H chain, YDF L chain | | Authors: | Meng, B, Yang, B, Wilson, I.A. | | Deposit date: | 2019-09-20 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Inhibitory antibodies identify unique sites of therapeutic vulnerability in rhinovirus and other enteroviruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6EPN
 
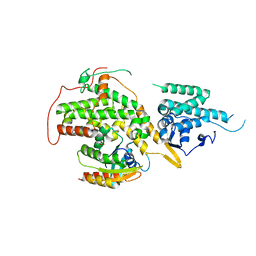 | | Ras guanine exchange factor SOS1 (Rem-cdc25) in complex with KRAS(G12C) and fragment screening hit F2 | | Descriptor: | 1-(3,4-dihydro-1~{H}-isoquinolin-2-yl)-2-oxidanyl-ethanone, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EPM
 
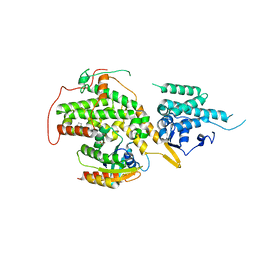 | | Ras guanine nucleotide exchange factor SOS1 (Rem-cdc25) in complex with KRAS(G12C) and fragment screening hit F1 | | Descriptor: | (1-phenyl-5,6-dihydro-4~{H}-cyclopenta[c]pyrazol-3-yl)methanamine, GLYCEROL, GTPase KRas, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EPP
 
 | | RAS GUANINE EXCHANGE FACTOR SOS1 (REM-CDC25) IN COMPLEX WITH KRAS(G12C) AND FRAGMENT SCREENING HIT F4 | | Descriptor: | GLYCEROL, GTPase KRas, Son of sevenless homolog 1, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6E8R
 
 | |
7LAW
 
 | | crystal structure of GITR complex with GITR-L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 18, Tumor necrosis factor receptor superfamily member 18 | | Authors: | Longenecker, K.L, Rogers, B, Bigelow, L, Judge, R.A, Alvarez, H. | | Deposit date: | 2021-01-07 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | An anti-PD-1-GITR-L bispecific agonist induces GITR clustering-mediated T cell activation for cancer immunotherapy.
Nat Cancer, 3, 2022
|
|
5ONI
 
 | | LOW-SALT STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA) IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,4-BUTANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, CHLORIDE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
5OOI
 
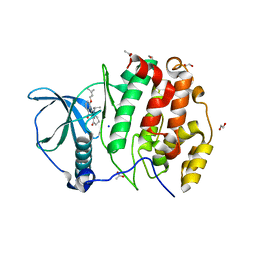 | | STRUCTURE OF PROTEIN KINASE CK2 CATALYTIC SUBUNIT (ISOFORM CK2ALPHA') IN COMPLEX WITH THE INDENOINDOLE-TYPE INHIBITOR 4P | | Descriptor: | 1,2-ETHANEDIOL, 4-(3-methylbut-2-enoxy)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, ACETATE ION, ... | | Authors: | Hochscherf, J, Lindenblatt, D, Witulski, B, Birus, R, Aichele, D, Marminon, C, Bouaziz, Z, Le Borgne, M, Jose, J, Niefind, K. | | Deposit date: | 2017-08-07 | | Release date: | 2017-12-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Unexpected Binding Mode of a Potent Indeno[1,2-b]indole-Type Inhibitor of Protein Kinase CK2 Revealed by Complex Structures with the Catalytic Subunit CK2 alpha and Its Paralog CK2 alpha '.
Pharmaceuticals (Basel), 10, 2017
|
|
8J3V
 
 | |
3KBP
 
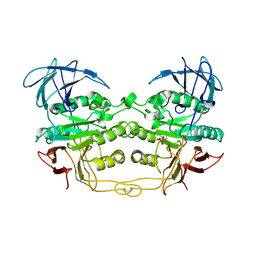 | | KIDNEY BEAN PURPLE ACID PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, PURPLE ACID PHOSPHATASE, ... | | Authors: | Klabunde, T, Strater, N, Krebs, B. | | Deposit date: | 1995-10-02 | | Release date: | 1996-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mechanism of Fe(III)-Zn(II) purple acid phosphatase based on crystal structures.
J.Mol.Biol., 259, 1996
|
|
5JAE
 
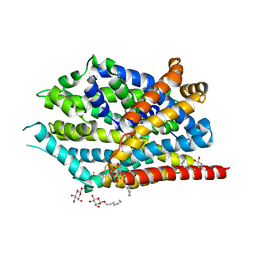 | | LeuT in the outward-oriented, Na+-free return state, P21 form at pH 6.5 | | Descriptor: | Transporter, octyl beta-D-glucopyranoside | | Authors: | Malinauskaite, L, Sahin, C, Said, S, Grouleff, J, Shahsavar, A, Bjerregaard, H, Noer, P, Severinsen, K, Boesen, T, Schiott, B, Sinning, S, Nissen, P. | | Deposit date: | 2016-04-12 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A conserved leucine occupies the empty substrate site of LeuT in the Na(+)-free return state.
Nat Commun, 7, 2016
|
|
6EPL
 
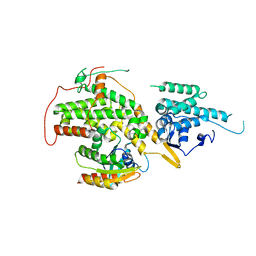 | | Ras guanine exchange factor SOS1 (Rem-cdc25) in complex with KRAS(G12C) | | Descriptor: | GLYCEROL, GTPase KRas, Son of sevenless homolog 1 | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6EPO
 
 | | RAS GUANINE EXCHANGE FACTOR SOS1 (REM-CDC25) IN COMPLEX WITH KRAS(G12C) AND FRAGMENT SCREENING HIT F3 | | Descriptor: | 3-(4-chlorophenyl)propan-1-amine, GLYCEROL, GTPase KRas, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Bader, B, Schroeder, J, Wortmann, L, Sautier, B, Kahmann, J, Wegener, D, Briem, H, Petersen, K, Badock, V. | | Deposit date: | 2017-10-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7MHF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHL
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 100 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
7MHM
 
 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
6LQI
 
 | | Cryo-EM structure of the mouse Piezo1 isoform Piezo1.1 | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Geng, J, Liu, W, Zhou, H, Zhang, T, Wang, L, Zhang, M, Shen, B, Li, X, Xiao, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | A Plug-and-Latch Mechanism for Gating the Mechanosensitive Piezo Channel.
Neuron, 106, 2020
|
|
5PAL
 
 | | CRYSTAL STRUCTURE OF THE UNIQUE PARVALBUMIN COMPONENT FROM MUSCLE OF THE LEOPARD SHARK (TRIAKIS SEMIFASCIATA). THE FIRST X-RAY STUDY OF AN ALPHA-PARVALBUMIN | | Descriptor: | CALCIUM ION, PARVALBUMIN | | Authors: | Roquet, F, Declercq, J.-P, Tinant, B, Rambaud, J, Parello, J. | | Deposit date: | 1991-09-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of the unique parvalbumin component from muscle of the leopard shark (Triakis semifasciata). The first X-ray study of an alpha-parvalbumin.
J.Mol.Biol., 223, 1992
|
|
7QCA
 
 | | Spraguea lophii ribosome | | Descriptor: | 40S Ribosomal protein S19, 40S ribosomal protein S0, 40S ribosomal protein S10, ... | | Authors: | Gil Diez, P, McLaren, M, Isupov, M.N, Daum, B, Conners, R, Williams, B. | | Deposit date: | 2021-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | CryoEM reveals that ribosomes in microsporidian spores are locked in a dimeric hibernating state
Nat Microbiol, 2023
|
|
2A2J
 
 | | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis | | Descriptor: | Pyridoxamine 5'-phosphate oxidase | | Authors: | Pedelacq, J.-D, Rho, B.-S, Kim, C.-Y, Waldo, G.S, Lekin, T.P, Segelke, B.W, Rupp, B, Hung, L.-W, Kim, S.-I, Terwilliger, T.C, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2005-06-22 | | Release date: | 2005-08-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a putative pyridoxine 5'-phosphate oxidase (Rv2607) from Mycobacterium tuberculosis.
Proteins, 62, 2005
|
|
1QVE
 
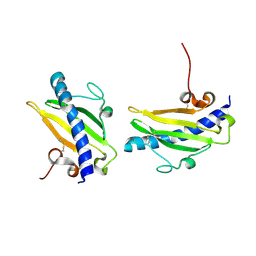 | |
6TW2
 
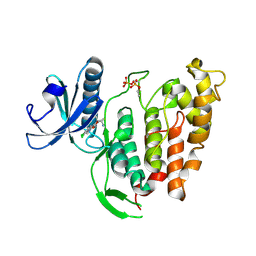 | | Re-refined crystal structure of di-phosphorylated human CLK1 in complex with a novel substituted indole inhibitor | | Descriptor: | Dual specificity protein kinase CLK1, ethyl 3-[(E)-2-amino-1-cyanoethenyl]-6,7-dichloro-1-methyl-1H-indole-2-carboxylate | | Authors: | Loll, B, Haltenhof, T, Heyd, F, Wahl, M.C. | | Deposit date: | 2020-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Kinase-Based Body-Temperature Sensor Globally Controls Alternative Splicing and Gene Expression.
Mol.Cell, 78, 2020
|
|
2ANJ
 
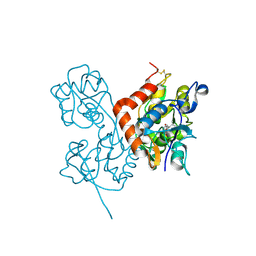 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
