4GF7
 
 | | Crystal structure of 2-Methyl-3-hydroxypyridine-5-carboxylic acid oxygenase (MHPCO), unliganded form | | Descriptor: | 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kobayashi, J, Yoshida, H, Mikami, B, Hayashi, H, Kamitori, S, Sawa, Y, Yagi, T. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase
To be Published
|
|
4UWM
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7DBH
 
 | | The mouse nucleosome structure containing H3mm18 | | Descriptor: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | Authors: | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | Deposit date: | 2020-10-20 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
3VPE
 
 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 | | Descriptor: | ACETATE ION, GLYCEROL, Metallo-beta-lactamase, ... | | Authors: | Wachino, J, Yamaguchi, Y, Mori, S, Arakawa, Y, Shibayama, K. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Subclass B3 Metallo-beta-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo- -Lactamase Inhibitor Mercaptoacetate
Antimicrob.Agents Chemother., 57, 2013
|
|
7DQ6
 
 | | Crystal structure of HitB in complex with (S)-beta-3-Br-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl N-[(3S)-3-azanyl-3-(3-bromophenyl)propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7DQ5
 
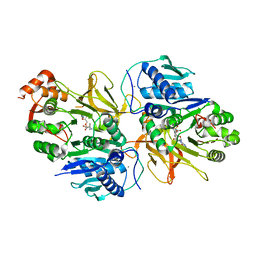 | | Crystal structure of HitB in complex with (S)-beta-phenylalanine sulfamoyladenosine | | Descriptor: | CALCIUM ION, Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-phenyl-propanoyl]sulfamate | | Authors: | Kudo, F, Takahashi, S, Miyanaga, A, Nakazawa, Y, Eguchi, T. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Mutational Biosynthesis of Hitachimycin Analogs Controlled by the beta-Amino Acid-Selective Adenylation Enzyme HitB.
Acs Chem.Biol., 16, 2021
|
|
7D0N
 
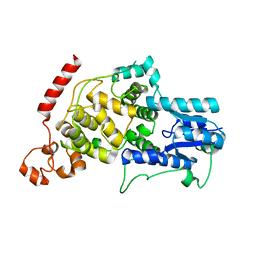 | | Crystal structure of mouse CRY2 apo form | | Descriptor: | Cryptochrome-2 | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-09-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7DLI
 
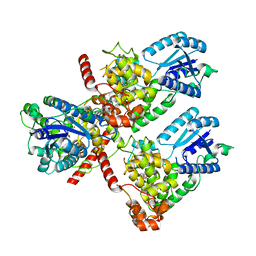 | | Crystal structure of mouse CRY1 in complex with KL001 compound | | Descriptor: | 1,2-ETHANEDIOL, Cryptochrome-1, N-[(2R)-3-carbazol-9-yl-2-oxidanyl-propyl]-N-(furan-2-ylmethyl)methanesulfonamide | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-11-27 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D0M
 
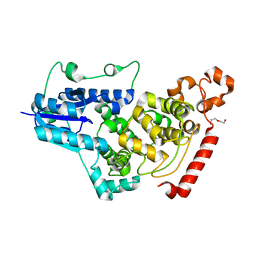 | | Crystal structure of mouse CRY1 with bound cryoprotectant | | Descriptor: | Cryptochrome-1, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL | | Authors: | Miller, S.A, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-09-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural differences in the FAD-binding pockets and lid loops of mammalian CRY1 and CRY2 for isoform-selective regulation.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6LUE
 
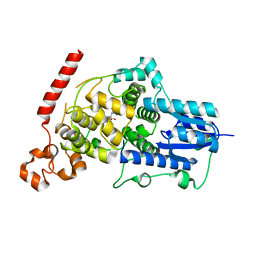 | | Crystal structure of mouse Cryptochrome 1 in complex with compound KL201 | | Descriptor: | 2-bromanyl-N-(5,6,7,8-tetrahydro-[1]benzothiolo[2,3-d]pyrimidin-4-yl)benzamide, Cryptochrome-1 | | Authors: | Miller, S, Aikawa, Y, Hirota, T. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An Isoform-Selective Modulator of Cryptochrome 1 Regulates Circadian Rhythms in Mammals.
Cell Chem Biol, 27, 2020
|
|
3VQZ
 
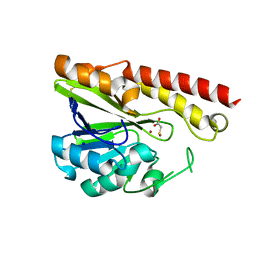 | | Crystal structure of metallo-beta-lactamase, SMB-1, in a complex with mercaptoacetic acid | | Descriptor: | Metallo-beta-lactamase, SODIUM ION, SULFANYLACETIC ACID, ... | | Authors: | Wachino, J, Yamaguchi, Y, Mori, S, Arakawa, Y, Shibayama, K. | | Deposit date: | 2012-04-02 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Subclass B3 Metallo-beta-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo- -Lactamase Inhibitor Mercaptoacetate
Antimicrob.Agents Chemother., 57, 2013
|
|
6LKR
 
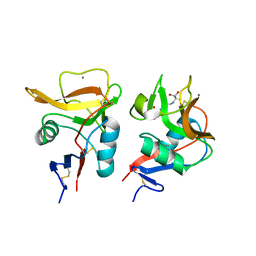 | | Crystal structure of mouse DCAR2 CRD domain complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
8IMD
 
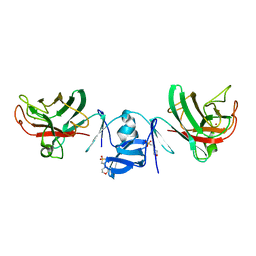 | | Crystal structure of Cu/Zn Superoxide dismutase from Paenibacillus lautus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cu/Zn-Superoxide dismutase, ... | | Authors: | Narikiyo, S, Furukawa, Y, Akutsu, M. | | Deposit date: | 2023-03-06 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of a novel cysteine-less Cu/Zn-superoxide dismutase in Paenibacillus lautus missing a conserved disulfide bond.
J.Biol.Chem., 299, 2023
|
|
3VGV
 
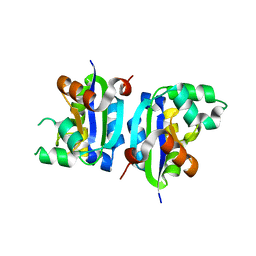 | | E134A mutant nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-21 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
4GHA
 
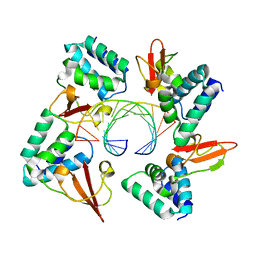 | | Crystal structure of Marburg virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor VP35, RNA (5'-R(*CP*UP*AP*GP*AP*CP*GP*UP*CP*UP*AP*G)-3') | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
4GH9
 
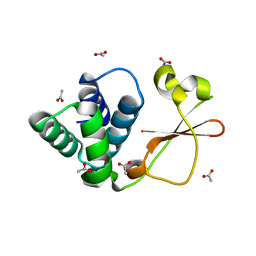 | | Crystal structure of Marburg virus VP35 RNA binding domain | | Descriptor: | ACETATE ION, Polymerase cofactor VP35 | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
7D20
 
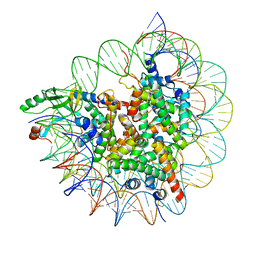 | | Cryo-EM structure of SET8-CENP-A-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7D1Z
 
 | | Cryo-EM structure of SET8-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7D69
 
 | | Cryo-EM structure of the nucleosome containing Giardia histones | | Descriptor: | 601L DNA (145-MER), Histone H2A, Histone H2B, ... | | Authors: | Sato, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structure of the nucleosome core particle containing Giardia lamblia histones.
Nucleic Acids Res., 49, 2021
|
|
8IHL
 
 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
8JLB
 
 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
3VGS
 
 | | Wild-type nucleoside diphosphate kinase derived from Halomonas sp. 593 | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Okazaki, N, Yonezawa, Y, Arai, S, Matsumoto, F, Tamada, T, Tokunaga, H, Ishibashi, M, Tokunaga, M, Kuroki, R. | | Deposit date: | 2011-08-20 | | Release date: | 2012-07-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural mechanism for dimeric to tetrameric oligomer conversion in Halomonas sp. nucleoside diphosphate kinase
Protein Sci., 21, 2012
|
|
6L2D
 
 | | Crystal structure of a cupin protein (tm1459) in copper (Cu) substituted form | | Descriptor: | COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.198 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L2E
 
 | | Crystal structure of a cupin protein (tm1459, H52A mutant) in copper (Cu) substituted form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Cupin_2 domain-containing protein | | Authors: | Fujieda, N, Ichihashi, H, Nishikawa, Y, Kurisu, G, Itoh, S. | | Deposit date: | 2019-10-03 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Cupin Variants as a Macromolecular Ligand Library for Stereoselective Michael Addition of Nitroalkanes.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8JL9
 
 | | Cryo-EM structure of the human nucleosome with scFv | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
