2ZNP
 
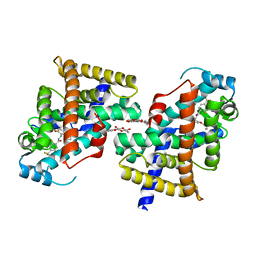 | | Human PPAR delta ligand binding domain in complex with a synthetic agonist TIPP204 | | Descriptor: | (2S)-2-{4-butoxy-3-[({[2-fluoro-4-(trifluoromethyl)phenyl]carbonyl}amino)methyl]benzyl}butanoic acid, Peroxisome proliferator-activated receptor delta, heptyl beta-D-glucopyranoside | | Authors: | Oyama, T, Hirakawa, Y, Nagasawa, N, Miyachi, H, Morikawa, K. | | Deposit date: | 2008-04-30 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Adaptability and selectivity of human peroxisome proliferator-activated receptor (PPAR) pan agonists revealed from crystal structures
Acta Crystallogr.,Sect.D, 65, 2009
|
|
5AXO
 
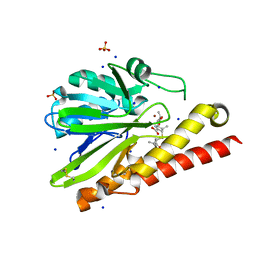 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Metallo-beta-lactamase, ... | | Authors: | Wachino, J, Arakawa, Y. | | Deposit date: | 2015-07-31 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Meropenem
To Be Published
|
|
5AYH
 
 | |
5B1U
 
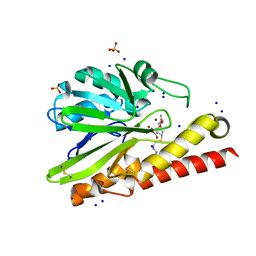 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, Metallo-beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Arakawa, Y. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal Structure of Metallo-beta-Lactamase SMB-1 Bound to Hydrolyzed Imipenem
To be published
|
|
2P6I
 
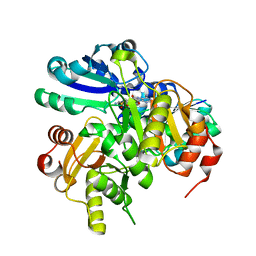 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Morikawa, Y, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-18 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
4FRV
 
 | | Crystal structure of mutated cyclophilin B that causes hyperelastosis cutis in the American Quarter Horse | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase, ... | | Authors: | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | Deposit date: | 2012-06-26 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of wild-type and mutated cyclophilin B that causes hyperelastosis cutis in the American quarter horse.
BMC Res Notes, 5, 2012
|
|
3W1Z
 
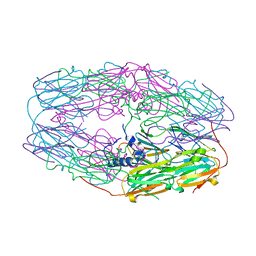 | | Heat shock protein 16.0 from Schizosaccharomyces pombe | | Descriptor: | Heat shock protein 16 | | Authors: | Hanazono, Y, Takeda, K, Akiyama, N, Aikawa, Y, Miki, K. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Nonequivalence Observed for the 16-Meric Structure of a Small Heat Shock Protein, SpHsp16.0, from Schizosaccharomyces pombe
Structure, 21, 2013
|
|
3VQI
 
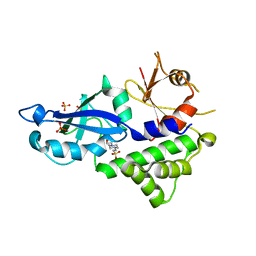 | | Crystal structure of Kluyveromyces marxianus Atg5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Atg5, SULFATE ION | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-03-24 | | Release date: | 2012-08-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
3WIN
 
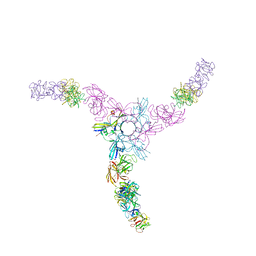 | | Clostridium botulinum Hemagglutinin | | Descriptor: | 17 kD hemagglutinin component, HA1, HA3 | | Authors: | Amatsu, S, Sugawara, Y, Matsumura, T, Fujinaga, Y, Kitadokoro, K. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structure of Clostridium botulinum Whole Hemagglutinin Reveals a Huge Triskelion-shaped Molecular Complex
J.Biol.Chem., 288, 2013
|
|
4FRU
 
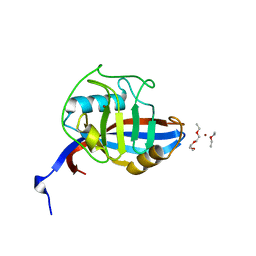 | | Crystal structure of horse wild-type cyclophilin B | | Descriptor: | 1-ETHOXY-2-(2-METHOXYETHOXY)ETHANE, DI(HYDROXYETHYL)ETHER, Peptidyl-prolyl cis-trans isomerase, ... | | Authors: | Boudko, S.P, Ishikawa, Y, Bachinger, H.P. | | Deposit date: | 2012-06-26 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of wild-type and mutated cyclophilin B that causes hyperelastosis cutis in the American quarter horse.
BMC Res Notes, 5, 2012
|
|
4GH9
 
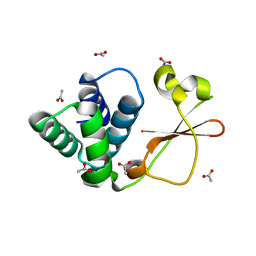 | | Crystal structure of Marburg virus VP35 RNA binding domain | | Descriptor: | ACETATE ION, Polymerase cofactor VP35 | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
7WWY
 
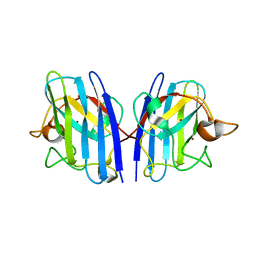 | |
7WWT
 
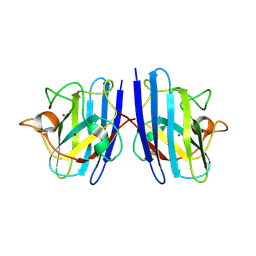 | | Cu/Zn-superoxide dismutase from dog (Canis familiaris) | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Narikiyo, S, Furukawa, Y, Akutsu, M. | | Deposit date: | 2022-02-14 | | Release date: | 2023-02-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Intrinsic structural vulnerability in the hydrophobic core induces species-specific aggregation of canine SOD1 with degenerative myelopathy-linked E40K mutation.
J.Biol.Chem., 299, 2023
|
|
7WX0
 
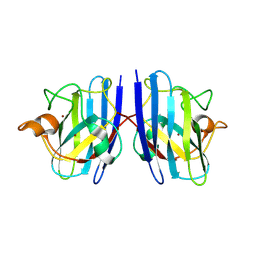 | |
7WX1
 
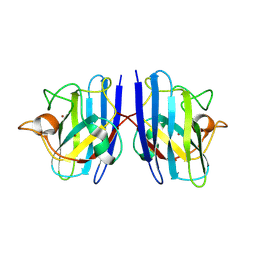 | |
2KFJ
 
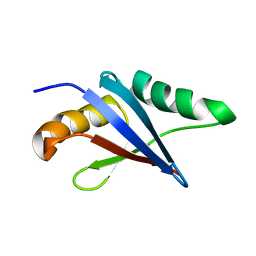 | | Solution structure of the loop deletion mutant of PB1 domain of Cdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Ogura, K, Tandai, T, Yoshinaga, S, Kobashigawa, Y, Kumeta, H, Inagaki, F. | | Deposit date: | 2009-02-22 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
4GHA
 
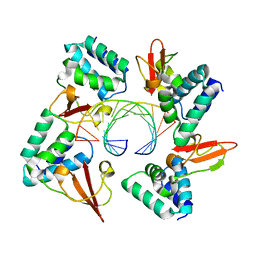 | | Crystal structure of Marburg virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor VP35, RNA (5'-R(*CP*UP*AP*GP*AP*CP*GP*UP*CP*UP*AP*G)-3') | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
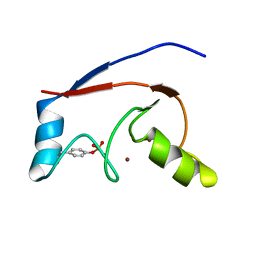
2LDR
 
 | |
3VQZ
 
 | | Crystal structure of metallo-beta-lactamase, SMB-1, in a complex with mercaptoacetic acid | | Descriptor: | Metallo-beta-lactamase, SODIUM ION, SULFANYLACETIC ACID, ... | | Authors: | Wachino, J, Yamaguchi, Y, Mori, S, Arakawa, Y, Shibayama, K. | | Deposit date: | 2012-04-02 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Subclass B3 Metallo-beta-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo- -Lactamase Inhibitor Mercaptoacetate
Antimicrob.Agents Chemother., 57, 2013
|
|
3AP2
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP,C4 peptide, and phosphate ion | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2: Insights into substrate-binding and catalysis of post-translational protein tyrosine sulfation
To be Published
|
|
5YXM
 
 | | Crystal structure of Chlamydomonas Outer Arm Dynein Light Chain 1 | | Descriptor: | Dynein light chain 1, axonemal, PHOSPHATE ION | | Authors: | Toda, A, Tanaka, H, Nishikawa, Y, Yagi, T, Kurisu, G. | | Deposit date: | 2017-12-06 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Structural atlas of dynein motors at atomic resolution.
Biophys Rev, 10, 2018
|
|
3V10
 
 | | Crystal structure of the collagen binding domain of Erysipelothrix rhusiopathiae surface protein RspB | | Descriptor: | Rhusiopathiae surface protein B | | Authors: | Ponnuraj, K, Swarmistha devi, A, Ogawa, Y, Shimoji, Y, Subramainan, B. | | Deposit date: | 2011-12-09 | | Release date: | 2012-10-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Collagen adhesin-nanoparticle interaction impairs adhesin's ligand binding mechanism
Biochim.Biophys.Acta, 1820, 2012
|
|
3VPE
 
 | | Crystal Structure of Metallo-beta-Lactamase SMB-1 | | Descriptor: | ACETATE ION, GLYCEROL, Metallo-beta-lactamase, ... | | Authors: | Wachino, J, Yamaguchi, Y, Mori, S, Arakawa, Y, Shibayama, K. | | Deposit date: | 2012-02-29 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Subclass B3 Metallo-beta-Lactamase SMB-1 and the Mode of Inhibition by the Common Metallo- -Lactamase Inhibitor Mercaptoacetate
Antimicrob.Agents Chemother., 57, 2013
|
|
3AP3
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Protein-tyrosine sulfotransferase 2 | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
3AP1
 
 | | Crystal structure of human tyrosylprotein sulfotransferase-2 complexed with PAP and C4 peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, C4 peptide, GLYCEROL, ... | | Authors: | Teramoto, T, Fujikawa, Y, Kawaguchi, Y, Kurogi, K, Soejima, M, Adachi, R, Nakanishi, Y, Mishiro-Sato, E, Liu, M.-C, Sakakibara, Y, Suiko, M, Kimura, M, Kakuta, Y. | | Deposit date: | 2010-10-09 | | Release date: | 2011-10-26 | | Last modified: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction.
Nat Commun, 4, 2013
|
|
