7O8G
 
 | | NmHR light state structure at 10 ps after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8O
 
 | | NmHR light state structure at 12.5 ms (10 - 15 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8U
 
 | | NmHR light state structure at 45 ms (40 - 50 ms) after photoexcitation determined by serial millisecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7U8K
 
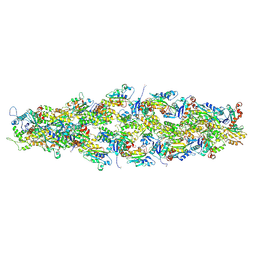 | | Magic Angle Spinning NMR Structure of Human Cofilin-2 Assembled on Actin Filaments | | Descriptor: | Actin, alpha skeletal muscle, Cofilin-2 | | Authors: | Kraus, J, Russell, R, Kudryashova, E, Xu, C, Katyal, N, Kudryashov, D, Perilla, J.R, Polenova, T. | | Deposit date: | 2022-03-08 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-23 | | Method: | SOLID-STATE NMR | | Cite: | Magic angle spinning NMR structure of human cofilin-2 assembled on actin filaments reveals isoform-specific conformation and binding mode.
Nat Commun, 13, 2022
|
|
8D3P
 
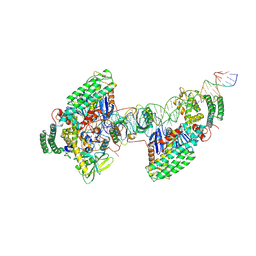 | | Type I-C Cas4-Cas1-Cas2 complex bound to half-site integration intermediate (HSI) | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
5XNT
 
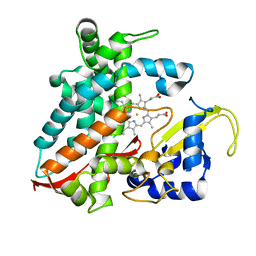 | | Structure of CYP106A2 from Bacillus sp. PAMC 23377 | | Descriptor: | Cytochrome P450 CYP106, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Kim, K.-H, Bikash, D, Park, S.-H, Park, H, Oh, T.-J, Lee, J.H. | | Deposit date: | 2017-05-24 | | Release date: | 2018-04-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and Functional Characterization of a Cytochrome P450 (BaCYP106A2) fromBacillussp. PAMC 23377.
J. Microbiol. Biotechnol., 27, 2017
|
|
7L33
 
 | | X-ray Structure of a Cu-Bound De Novo Designed Peptide Trimer | | Descriptor: | COPPER (II) ION, Cu-3SCC | | Authors: | Chakraborty, S, Wawrzak, Z, Prasad, P, Mitra, S, Prakash, D. | | Deposit date: | 2020-12-17 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | De Novo Design of a Self-Assembled Artificial Copper Peptide that Activates and Reduces Peroxide
Acs Catalysis, 11, 2021
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
5L8Z
 
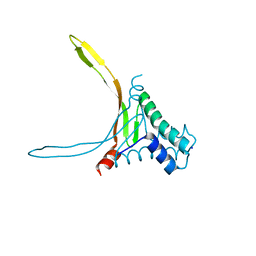 | | Structure of thermostable DNA-binding HU protein from micoplasma Spiroplasma melliferum | | Descriptor: | DNA-binding protein, SODIUM ION | | Authors: | Boyko, K.M, Gorbacheva, M.A, Rakitina, T.V, Korzhenevskiy, D.A, Kamashev, D.E, Vanyushkina, A.A, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2016-06-09 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the high thermal stability of the histone-like HU protein from the mollicute Spiroplasma melliferum KC3.
Sci Rep, 6, 2016
|
|
8D3M
 
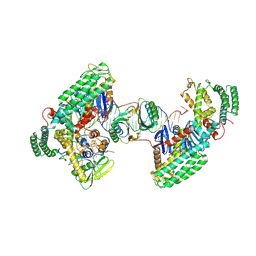 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/Processed prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3L
 
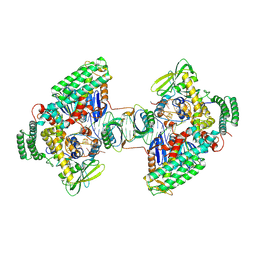 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/PAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
8D3Q
 
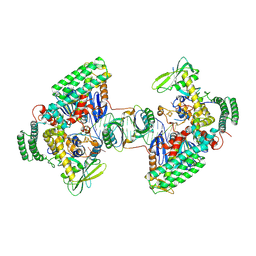 | | Type I-C Cas4-Cas1-Cas2 complex bound to a PAM/NoPAM prespacer | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endonuclease Cas2, CRISPR-associated exonuclease Cas4, ... | | Authors: | Dhingra, Y, Suresh, S.K, Juneja, P, Sashital, D.G. | | Deposit date: | 2022-06-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | PAM binding ensures orientational integration during Cas4-Cas1-Cas2-mediated CRISPR adaptation.
Mol.Cell, 82, 2022
|
|
5IVU
 
 | |
2Q36
 
 | | Actin Dimer Cross-linked between Residues 191 and 374 and complexed with Kabiramide C | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Pashkov, I, Kudryashov, D.S, Reisler, E, Yeates, T.O. | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiple crystal structures of actin dimers and their implications for interactions in the actin filament.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
2Q1N
 
 | | Actin Dimer Cross-linked Between Residues 41 and 374 | | Descriptor: | Actin, alpha skeletal muscle, CALCIUM ION, ... | | Authors: | Sawaya, M.R, Pashkov, I, Kudryashov, D.S, Adisetiyo, H, Reisler, E, Yeates, T.O. | | Deposit date: | 2007-05-25 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Multiple crystal structures of actin dimers and their implications for interactions in the actin filament.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
1ENU
 
 | | A new target for shigellosis: Rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase | | Descriptor: | 4-AMINOPHTHALHYDRAZIDE, TRNA-GUANINE TRANSGLYCOSYLASE, ZINC ION | | Authors: | Gradler, U, Gerber, H.D, Goodenough-Lashua, D.M, Garcia, G.A, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
1F3E
 
 | | A NEW TARGET FOR SHIGELLOSIS: RATIONAL DESIGN AND CRYSTALLOGRAPHIC STUDIES OF INHIBITORS OF TRNA-GUANINE TRANSGLYCOSYLASE | | Descriptor: | 3,5-DIAMINOPHTHALHYDRAZIDE, QUEUINE TRNA-RIBOSYLTRANSFERASE, ZINC ION | | Authors: | Graedler, U, Gerber, H.-D, Goodenough-Lashua, D.M, Garcia, G.A.G, Ficner, R, Reuter, K, Stubbs, M.T, Klebe, G. | | Deposit date: | 2000-06-02 | | Release date: | 2000-06-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A new target for shigellosis: rational design and crystallographic studies of inhibitors of tRNA-guanine transglycosylase.
J.Mol.Biol., 306, 2001
|
|
7YEB
 
 | | TR-SFX MmCPDII-DNA complex: 3.35 ns snapshot. Includes 3.35 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photolyase, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEI
 
 | | TR-SFX MmCPDII-DNA complex: 10 ns time-point collected in SACLA. Includes 10 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEC
 
 | | TR-SFX MmCPDII-DNA complex: 6 ns snapshot. Includes 6 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA, Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEK
 
 | | TR-SFX MmCPDII-DNA complex: 500 ns time-point collected in SACLA. Includes 500 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEJ
 
 | | TR-SFX MmCPDII-DNA complex: 100 ns time-point collected in SACLA. Includes 100 ns, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEL
 
 | | TR-SFX MmCPDII-DNA complex: 25 us time-point collected in SACLA. Includes 25 us, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7YEM
 
 | | TR-SFX MmCPDII-DNA complex: 200 us time-point collected in SACLA. Includes 200 us, dark, and extrapolated structure factors | | Descriptor: | CPD photolesion containing DNA after repair, Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
