4ZKK
 
 | | The novel double-fold structure of d(GCATGCATGC) | | Descriptor: | COBALT (II) ION, DNA (5'-D(*GP*CP*AP*TP*GP*CP*AP*TP*GP*C)-3') | | Authors: | Thirugnanasambandam, A, Karthik, S, Mandal, P.K, Gautham, N. | | Deposit date: | 2015-04-30 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The novel double-folded structure of d(GCATGCATGC): a possible model for triplet-repeat sequences
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5FHL
 
 | |
5FHJ
 
 | |
1XJU
 
 | |
1XJT
 
 | |
6ALK
 
 | | NMR solution structure of the major beech pollen allergen Fag s 1 | | Descriptor: | Fag s 1 pollen allergen | | Authors: | Moraes, A.H, Asam, A, Almeida, F.C.L, Wallner, M, Ferreira, F, Valente, A.P. | | Deposit date: | 2017-08-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for cross-reactivity and conformation fluctuation of the major beech pollen allergen Fag s 1.
Sci Rep, 8, 2018
|
|
4ZPU
 
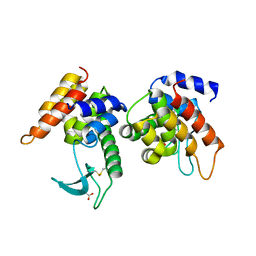 | |
5IYE
 
 | | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 resolution | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*GP*TP*AP*CP*CP*CP*GP*GP*G)-3'), ZINC ION | | Authors: | Karthik, S, Thirugnanasambandam, A, Mandal, P.K, Gautham, N. | | Deposit date: | 2016-03-24 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Comparison of X-ray crystal structures of a tetradecamer sequence d(CCCGGGTACCCGGG)2 at 1.7 angstrom resolution.
Nucleosides Nucleotides Nucleic Acids, 36, 2017
|
|
5IYG
 
 | |
5IYJ
 
 | |
3HDE
 
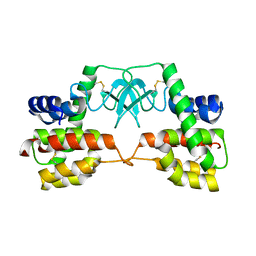 | | Crystal structure of full-length endolysin R21 from phage 21 | | Descriptor: | Lysozyme | | Authors: | Sun, Q, Arockiasamy, A, McKee, E, Caronna, E, Sacchettini, J.C. | | Deposit date: | 2009-05-07 | | Release date: | 2009-11-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Regulation of a muralytic enzyme by dynamic membrane topology.
Nat.Struct.Mol.Biol., 16, 2009
|
|
5IQY
 
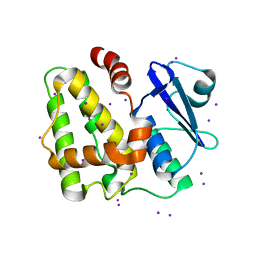 | | Structure of apo-Dehydroascorbate Reductase from Pennisetum Glaucum phased by Iodide-SAD method | | Descriptor: | Dehydroascorbate reductase, IODIDE ION | | Authors: | Das, B.K, Kumar, A, Manidola, P, Arockiasamy, A. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase
Biochem.Biophys.Res.Commun., 473, 2016
|
|
6A1Q
 
 | | Crystal structures of disordered Z-type helices | | Descriptor: | DNA (5'-D(P*CP*AP*CP*A)-3'), DNA (5'-D(P*TP*GP*TP*G)-3') | | Authors: | Karthik, S, Mandal, P.K, Thirugnanasambandam, A, Gautham, N. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of disordered Z-type helices.
Nucleosides Nucleotides Nucleic Acids, 38, 2019
|
|
5WV7
 
 | |
3NSG
 
 | | Crystal Structure of OmpF, an Outer Membrane Protein from Salmonella typhi | | Descriptor: | CITRATE ANION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Balasubramaniam, D, Arockiasamy, A, Sharma, A, Krishnaswamy, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Asymmetric pore occupancy in crystal structure of OmpF porin from Salmonella typhi
J.Struct.Biol., 178, 2012
|
|
5EVO
 
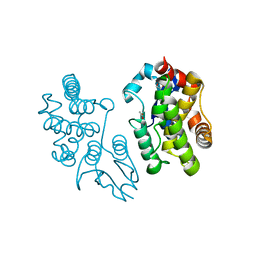 | | Structure of Dehydroascrobate Reductase from Pennisetum Americanum in complex with two non-native ligands, Acetate in the G-site and Glycerol in the H-site | | Descriptor: | ACETATE ION, Dehydroascorbate reductase, GLYCEROL | | Authors: | Kumar, A.O, Das, B.K, Arockiasamy, A. | | Deposit date: | 2015-11-20 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase.
Biochem.Biophys.Res.Commun., 473, 2016
|
|
7DK1
 
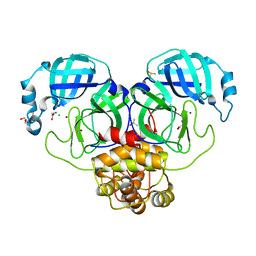 | | Crystal structure of Zinc bound SARS-CoV-2 main protease | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Sonkar, K.S, Panchariya, L, Kuila, S, Khan, W.A, Arockiasamy, A. | | Deposit date: | 2020-11-22 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Zinc 2+ ion inhibits SARS-CoV-2 main protease and viral replication in vitro.
Chem.Commun.(Camb.), 57, 2021
|
|
7W5D
 
 | |
6A1O
 
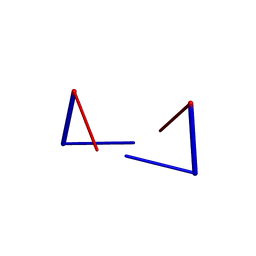 | | Crystal structures of disordered Z-type helices | | Descriptor: | DNA (5'-D(P*CP*A)-3'), DNA (5'-D(P*CP*AP*CP*A)-3'), DNA (5'-D(P*TP*G)-3'), ... | | Authors: | Karthik, S, Mandal, P.K, Thirugnanasambandam, A, Gautham, N. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structures of disordered Z-type helices.
Nucleosides Nucleotides Nucleic Acids, 38, 2019
|
|
3OSX
 
 | |
1NKT
 
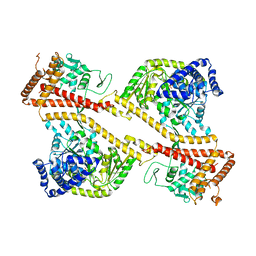 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS COMPLEX WITH ADPBS | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Preprotein translocase secA 1 subunit | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-03 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NL3
 
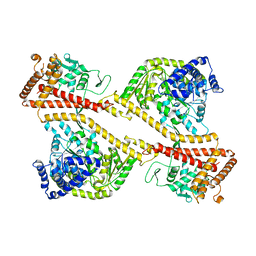 | | CRYSTAL STRUCTURE OF THE SECA PROTEIN TRANSLOCATION ATPASE FROM MYCOBACTERIUM TUBERCULOSIS in APO FORM | | Descriptor: | PREPROTEIN TRANSLOCASE SECA 1 SUBUNIT | | Authors: | Sharma, V, Arockiasamy, A, Ronning, D.R, Savva, C.G, Holzenburg, A, Braunstein, M, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of M. tuberculosis SecA, A Preprotein Translocating ATPase
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7XMP
 
 | |
7D9L
 
 | |
7DKP
 
 | |
