5UM9
 
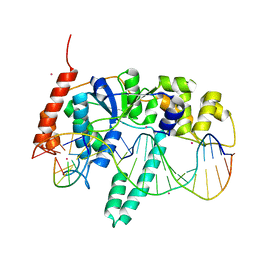 | | Flap endonuclease 1 (FEN1) D86N with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
2UGI
 
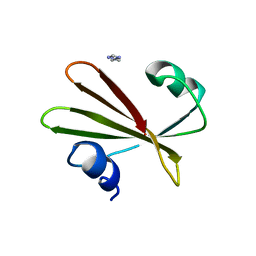 | | PROTEIN MIMICRY OF DNA FROM CRYSTAL STRUCTURES OF THE URACIL GLYCOSYLASE INHIBITOR PROTEIN AND ITS COMPLEX WITH ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | Descriptor: | IMIDAZOLE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-11-06 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2UUG
 
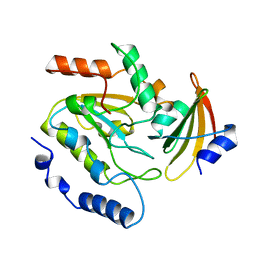 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH H187D MUTANT UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2W36
 
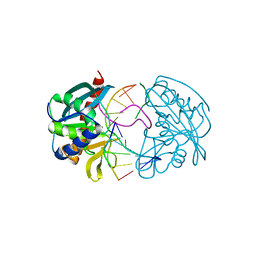 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W35
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2FZL
 
 | |
2FWR
 
 | | Structure of Archaeoglobus Fulgidis XPB | | Descriptor: | DNA repair protein RAD25, ISOPROPYL ALCOHOL, PHOSPHATE ION | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2006-02-02 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved XPB Core Structure and Motifs for DNA Unwinding: Implications for Pathway Selection of Transcription or Excision Repair
Mol.Cell, 22, 2006
|
|
2HI2
 
 | | Crystal structure of native Neisseria gonorrhoeae Type IV pilin at 2.3 Angstroms Resolution | | Descriptor: | Fimbrial protein, HEPTANE-1,2,3-TRIOL, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | Authors: | Craig, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions.
Mol.Cell, 23, 2006
|
|
1JWK
 
 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457A Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
1JWJ
 
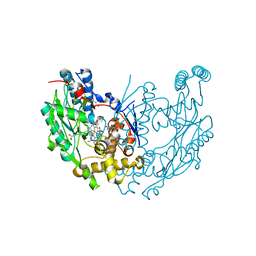 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457F Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
1KEA
 
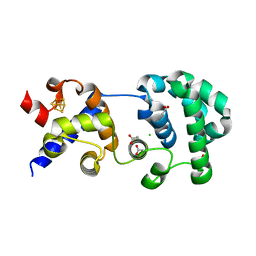 | | STRUCTURE OF A THERMOSTABLE THYMINE-DNA GLYCOSYLASE | | Descriptor: | ACETATE ION, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Mol, C.D, Arvai, A.S, Begley, T.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 2001-11-14 | | Release date: | 2002-01-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of a thermostable thymine-DNA glycosylase: evidence for base twisting to remove mismatched normal DNA bases.
J.Mol.Biol., 315, 2002
|
|
1WEG
 
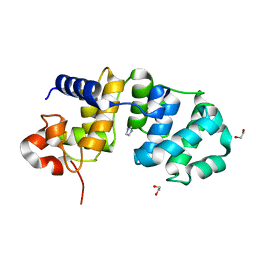 | | Catalytic Domain Of Muty From Escherichia Coli K142A Mutant | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, IMIDAZOLE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
1WWJ
 
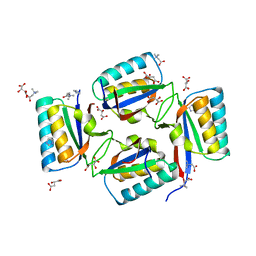 | | crystal structure of KaiB from Synechocystis sp. | | Descriptor: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | Authors: | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
1WEF
 
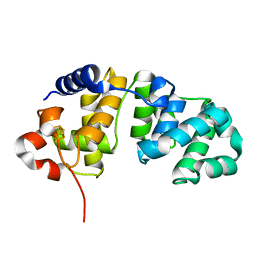 | |
1WEI
 
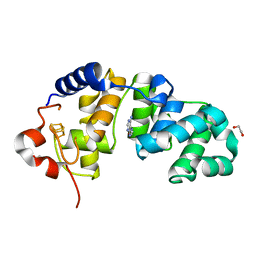 | | Catalytic Domain Of Muty From Escherichia Coli K20A Mutant Complexed To Adenine | | Descriptor: | 1,2-ETHANEDIOL, A/G-specific adenine glycosylase, ADENINE, ... | | Authors: | Hitomi, K, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-09-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reaction intermediates in the catalytic mechanism of Escherichia coli MutY DNA glycosylase
J.Biol.Chem., 279, 2004
|
|
1QOM
 
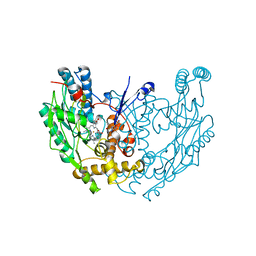 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH SWAPPED N-TERMINAL HOOK | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Crane, B.R, Rosenfeld, R.A, Arvai, A.S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-15 | | Release date: | 1999-12-15 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-Terminal Domain Swapping and Metal Ion Binding in Nitric Oxide Synthase Dimerization
Embo J., 18, 1999
|
|
1UGI
 
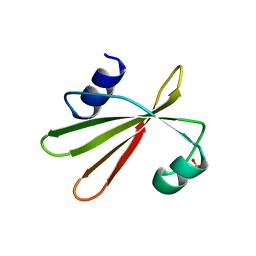 | | URACIL-DNA GLYCOSYLASE INHIBITOR PROTEIN | | Descriptor: | IMIDAZOLE, SULFATE ION, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-11-04 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
1VKG
 
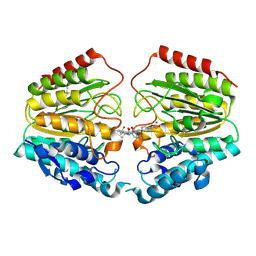 | | Crystal Structure of Human HDAC8 complexed with CRA-19156 | | Descriptor: | 5-(4-METHYL-BENZOYLAMINO)-BIPHENYL-3,4'-DICARBOXYLIC ACID 3-DIMETHYLAMIDE-4'-HYDROXYAMIDE, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-13 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1XBS
 
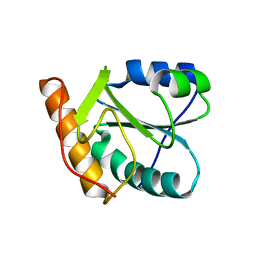 | | Crystal structure of human dim2: a dim1-like protein | | Descriptor: | Dim1-like protein | | Authors: | Simeoni, F, Arvai, A, Hopfner, K.-P, Bello, P, Gondeau, C, Heitz, F, Tainer, J, Divita, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Characterization and Crystal Structure of a Dim1 Family Associated Protein: Dim2
Biochemistry, 44, 2005
|
|
1UUG
 
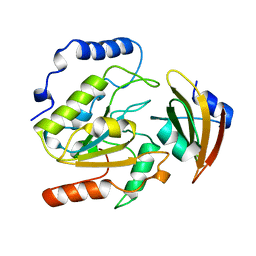 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH WILD-TYPE UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Mol, C.D, Arvai, A.S, Putnam, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
6VBH
 
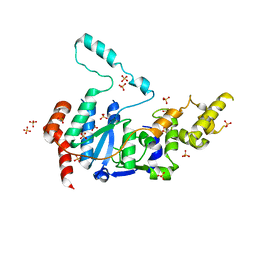 | | Human XPG endonuclease catalytic domain | | Descriptor: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VBA
 
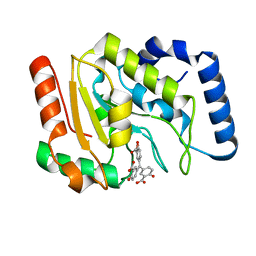 | | Structure of human Uracil DNA Glycosylase (UDG) bound to Aurintricarboxylic acid (ATA) | | Descriptor: | 3,3'-[(3-carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(6-hydroxybenzoic acid), Uracil-DNA glycosylase | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An effective human uracil-DNA glycosylase inhibitor targets the open pre-catalytic active site conformation.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
1T69
 
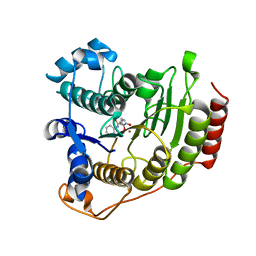 | | Crystal Structure of human HDAC8 complexed with SAHA | | Descriptor: | Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ZINC ION | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1T67
 
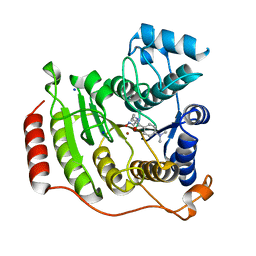 | | Crystal Structure of Human HDAC8 complexed with MS-344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
3SOJ
 
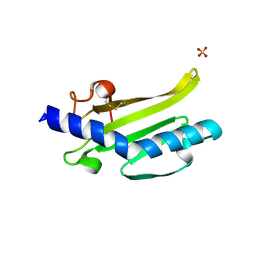 | | Francisella tularensis pilin PilE | | Descriptor: | PilE, SULFATE ION | | Authors: | Wood, T, Arvai, A.S, Shin, D.S, Hartung, S, Kolappan, S, Craig, L, Tainer, J.A. | | Deposit date: | 2011-06-30 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ultrahigh Resolution and Full-length Pilin Structures with Insights for Filament Assembly, Pathogenic Functions, and Vaccine Potential.
J.Biol.Chem., 286, 2011
|
|
