6VBA
 
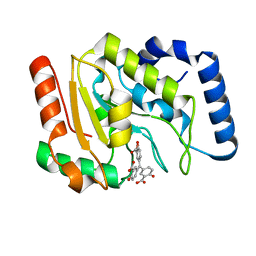 | | Structure of human Uracil DNA Glycosylase (UDG) bound to Aurintricarboxylic acid (ATA) | | Descriptor: | 3,3'-[(3-carboxy-4-oxocyclohexa-2,5-dien-1-ylidene)methylene]bis(6-hydroxybenzoic acid), Uracil-DNA glycosylase | | Authors: | Moiani, D, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An effective human uracil-DNA glycosylase inhibitor targets the open pre-catalytic active site conformation.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
4YDS
 
 | |
4AXN
 
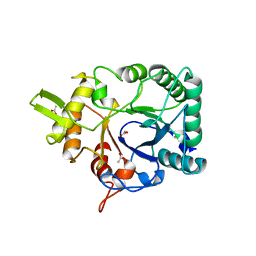 | | Hallmarks of processive and non-processive glycoside hydrolases revealed from computational and crystallographic studies of the Serratia marcescens chitinases | | Descriptor: | ACETATE ION, CALCIUM ION, CHITINASE C1 | | Authors: | Payne, C.M, Baban, J, Synstad, B, Backe, P.H, Arvai, A.S, Dalhus, B, Bjoras, M, Eijsink, V.G.H, Sorlie, M, Beckham, G.T, Vaaje-Kolstad, G. | | Deposit date: | 2012-06-13 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Hallmarks of Processivity in Glycoside Hydrolases from Crystallographic and Computational Studies of the Serratia Marcescens Chitinases.
J.Biol.Chem., 287, 2012
|
|
6VBH
 
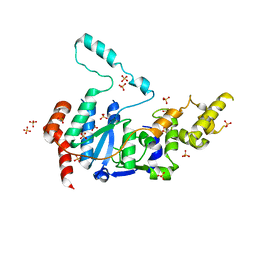 | | Human XPG endonuclease catalytic domain | | Descriptor: | DNA repair protein complementing XP-G cells,Flap endonuclease 1, SULFATE ION | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2019-12-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Human XPG nuclease structure, assembly, and activities with insights for neurodegeneration and cancer from pathogenic mutations.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1AV8
 
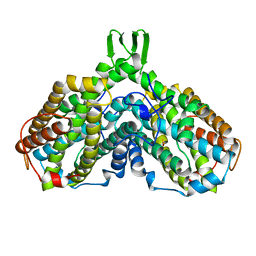 | | RIBONUCLEOTIDE REDUCTASE R2 SUBUNIT FROM E. COLI | | Descriptor: | MU-OXO-DIIRON, RIBONUCLEOTIDE REDUCTASE R2 | | Authors: | Han, S, Arvai, A, Tainer, J.A. | | Deposit date: | 1997-09-30 | | Release date: | 1998-10-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Characterization of Y122F R2 of Escherichia coli ribonucleotide reductase by time-resolved physical biochemical methods and X-ray crystallography.
Biochemistry, 37, 1998
|
|
3SOJ
 
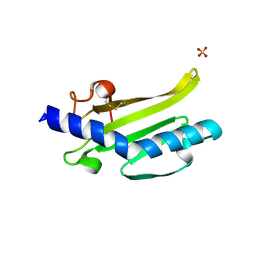 | | Francisella tularensis pilin PilE | | Descriptor: | PilE, SULFATE ION | | Authors: | Wood, T, Arvai, A.S, Shin, D.S, Hartung, S, Kolappan, S, Craig, L, Tainer, J.A. | | Deposit date: | 2011-06-30 | | Release date: | 2011-11-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Ultrahigh Resolution and Full-length Pilin Structures with Insights for Filament Assembly, Pathogenic Functions, and Vaccine Potential.
J.Biol.Chem., 286, 2011
|
|
1UGH
 
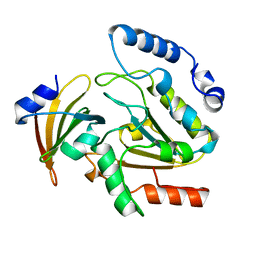 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | Descriptor: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | Authors: | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | Deposit date: | 1999-02-05 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
1CKS
 
 | |
2W36
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*TP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*BRUP*AP*CP*IP*GP*AP*BRUP*CP*GP)-3', ENDONUCLEASE V | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W35
 
 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2NOD
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND WATER BOUND IN ACTIVE CENTER | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-05 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
2UGI
 
 | | PROTEIN MIMICRY OF DNA FROM CRYSTAL STRUCTURES OF THE URACIL GLYCOSYLASE INHIBITOR PROTEIN AND ITS COMPLEX WITH ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE | | Descriptor: | IMIDAZOLE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-11-06 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2UUG
 
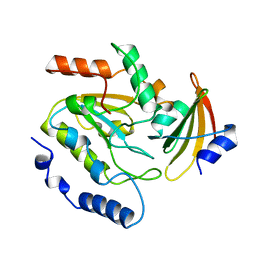 | | ESCHERICHIA COLI URACIL-DNA GLYCOSYLASE:INHIBITOR COMPLEX WITH H187D MUTANT UDG AND WILD-TYPE UGI | | Descriptor: | URACIL-DNA GLYCOSYLASE, URACIL-DNA GLYCOSYLASE INHIBITOR | | Authors: | Putnam, C.D, Arvai, A.S, Mol, C.D, Tainer, J.A. | | Deposit date: | 1998-10-31 | | Release date: | 1999-03-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase
J.Mol.Biol., 287, 1999
|
|
2FZL
 
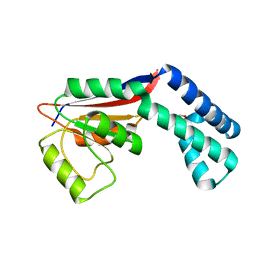 | |
2HI2
 
 | | Crystal structure of native Neisseria gonorrhoeae Type IV pilin at 2.3 Angstroms Resolution | | Descriptor: | Fimbrial protein, HEPTANE-1,2,3-TRIOL, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | Authors: | Craig, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Type IV Pilus Structure by Cryo-Electron Microscopy and Crystallography: Implications for Pilus Assembly and Functions.
Mol.Cell, 23, 2006
|
|
2FWR
 
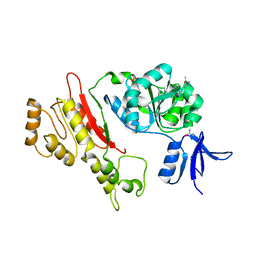 | | Structure of Archaeoglobus Fulgidis XPB | | Descriptor: | DNA repair protein RAD25, ISOPROPYL ALCOHOL, PHOSPHATE ION | | Authors: | Fan, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2006-02-02 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conserved XPB Core Structure and Motifs for DNA Unwinding: Implications for Pathway Selection of Transcription or Excision Repair
Mol.Cell, 22, 2006
|
|
1JWK
 
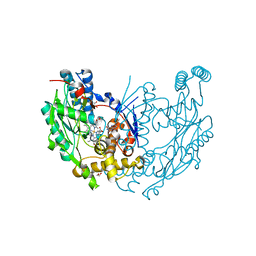 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457A Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 7,8-DIHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
3HRV
 
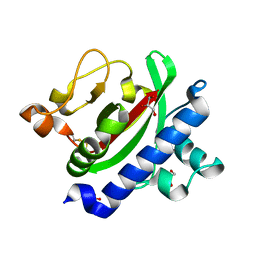 | | Crystal structure of TcpA, a Type IV pilin from Vibrio cholerae El Tor biotype | | Descriptor: | GLYCEROL, SULFATE ION, Toxin coregulated pilin | | Authors: | Craig, L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation.
Mol.Microbiol., 77, 2010
|
|
1XBS
 
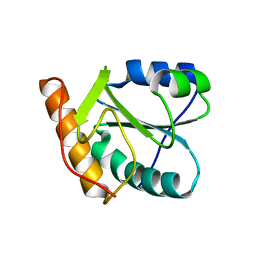 | | Crystal structure of human dim2: a dim1-like protein | | Descriptor: | Dim1-like protein | | Authors: | Simeoni, F, Arvai, A, Hopfner, K.-P, Bello, P, Gondeau, C, Heitz, F, Tainer, J, Divita, G. | | Deposit date: | 2004-08-31 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biochemical Characterization and Crystal Structure of a Dim1 Family Associated Protein: Dim2
Biochemistry, 44, 2005
|
|
1JWJ
 
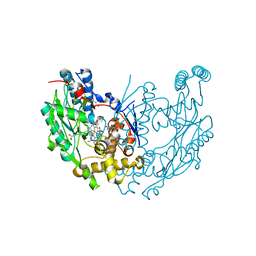 | | Murine Inducible Nitric Oxide Synthase Oxygenase Dimer (Delta 65) with W457F Mutation at Tetrahydrobiopterin Binding Site | | Descriptor: | 1,2-ETHANEDIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Aoyagi, M, Arvai, A.S, Ghosh, S, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2001-09-04 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of tetrahydrobiopterin binding-site mutants of inducible nitric oxide synthase oxygenase dimer and implicated roles of Trp457.
Biochemistry, 40, 2001
|
|
3QKR
 
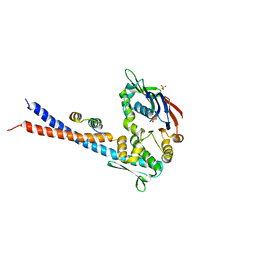 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, PHOSPHATE ION | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3GVA
 
 | |
3GX4
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with DNA | | Descriptor: | Alkyltransferase-like protein 1, COBALT HEXAMMINE(III), DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2009-04-01 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
1WWJ
 
 | | crystal structure of KaiB from Synechocystis sp. | | Descriptor: | Circadian clock protein kaiB, D-MALATE, IMIDAZOLE, ... | | Authors: | Hitomi, K, Oyama, T, Han, S, Arvai, A.S, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm.
J.Biol.Chem., 280, 2005
|
|
3GYH
 
 | | Crystal Structure Analysis of S. Pombe ATL in complex with damaged DNA containing POB | | Descriptor: | 1-PYRIDIN-3-YLBUTAN-1-ONE, Alkyltransferase-like protein 1, DNA (5'-D(*CP*TP*AP*CP*TP*AP*GP*CP*CP*AP*TP*GP*G)-3'), ... | | Authors: | Tubbs, J.L, Arvai, A.S, Tainer, J.A, Shin, D.S. | | Deposit date: | 2009-04-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Flipping of alkylated DNA damage bridges base and nucleotide excision repair.
Nature, 459, 2009
|
|
