4RHV
 
 | |
1VRH
 
 | | HRV14/SDZ 880-061 COMPLEX | | Descriptor: | 2-[4-(2H-1,4-BENZOTHIAZINE-3-YL)-PIPERAZINE-1-LY]-1,3-THIAZOLE-4-CARBOXYLIC ACID ETHYLESTER, RHINOVIRUS 14 | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1996-02-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and activity of piperazine-containing antirhinoviral agents and crystal structure of SDZ 880-061 bound to human rhinovirus 14.
J.Mol.Biol., 259, 1996
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8503 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4PUO
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*UP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-13 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
2RHK
 
 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | Authors: | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4W9S
 
 | | 2-(4-(1H-tetrazol-5-yl)phenyl)-5-hydroxypyrimidin-4(3H)-one bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 5-hydroxy-2-[4-(1H-tetrazol-5-yl)phenyl]pyrimidin-4(3H)-one, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phenyl Substituted 4-Hydroxypyridazin-3(2H)-ones and 5-Hydroxypyrimidin-4(3H)-ones: Inhibitors of Influenza A Endonuclease.
J.Med.Chem., 57, 2014
|
|
5I3U
 
 | |
8FE8
 
 | | Crystal Structure of HIV-1 RT in Complex with the non-nucleoside inhibitor 18b1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}pyrimidin-2-yl)amino]-2-[4-(methanesulfonyl)piperazin-1-yl]benzonitrile, Reverse transcriptase p51, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of diarylpyrimidine derivatives bearing piperazine sulfonyl as potent HIV-1 nonnucleoside reverse transcriptase inhibitors.
Commun Chem, 6, 2023
|
|
1HRI
 
 | | STRUCTURE DETERMINATION OF ANTIVIRAL COMPOUND SCH 38057 COMPLEXED WITH HUMAN RHINOVIRUS 14 | | Descriptor: | 1-[6-(2-CHLORO-4-METHYXYPHENOXY)-HEXYL]-IMIDAZOLE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Zhang, A, Nanni, R.G, Oren, D.A, Arnold, E. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure determination of antiviral compound SCH 38057 complexed with human rhinovirus 14.
J.Mol.Biol., 230, 1993
|
|
1HRV
 
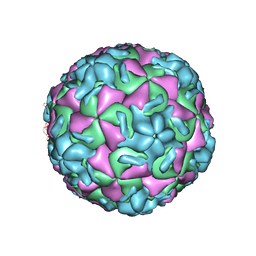 | | HRV14/SDZ 35-682 COMPLEX | | Descriptor: | 1-[2-HYDROXY-3-(4-CYCLOHEXYL-PHENOXY)-PROPYL]-4-(2-PYRIDYL)-PIPERAZINE, HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), ... | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1995-03-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | SDZ 35-682, a new picornavirus capsid-binding agent with potent antiviral activity.
Antiviral Res., 26, 1995
|
|
5HP1
 
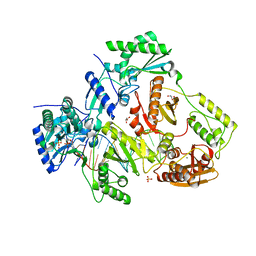 | |
5I42
 
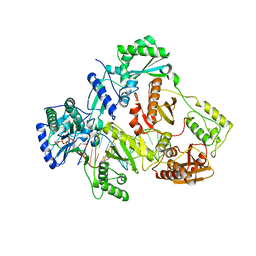 | | Structure of HIV-1 Reverse Transcriptase in complex with a DNA aptamer, AZTTP, and CA(2+) ion | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (38-MER), ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.
Acs Chem.Biol., 11, 2016
|
|
8FFX
 
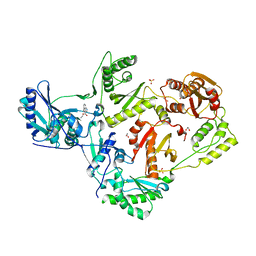 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 19980 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Reverse transcriptase/ribonuclease H, ... | | Authors: | Rumrill, S.R, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Targeting HIV-1 Reverse Transcriptase Using a Fragment-Based Approach.
Molecules, 28, 2023
|
|
2BE2
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with R221239 | | Descriptor: | 4-(3,5-DIMETHYLPHENOXY)-5-(FURAN-2-YLMETHYLSULFANYLMETHYL)-3-IODO-6-METHYLPYRIDIN-2(1H)-ONE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Himmel, D.M, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Nguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
2B5J
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R165481 | | Descriptor: | (2E)-3-{3-[(5-ETHYL-3-IODO-6-METHYL-2-OXO-1,2-DIHYDROPYRIDIN-4-YL)OXY]PHENYL}ACRYLONITRILE, MANGANESE (II) ION, Reverse transcriptase P51 SUBUNIT, ... | | Authors: | Himmel, D.H, Das, K, Clark Jr, A.D, Hughes, S.H, Benjahad, A, Oumouch, S, Guillemont, J, Coupa, S, Poncelet, A, Csoka, I, Meyer, C, Andries, K, Mguyen, C.H, Grierson, D.S, Arnold, E. | | Deposit date: | 2005-09-28 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
2BAN
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with JANSSEN-R157208 | | Descriptor: | 5-ETHYL-3-[(2-METHOXYETHYL)METHYLAMINO]-6-METHYL-4-(3-METHYLBENZYL)PYRIDIN-2(1H)-ONE, MANGANESE (II) ION, Reverse transcriptase P51 subunit, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2005-10-14 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structures for HIV-1 Reverse Transcriptase in Complexes with Three Pyridinone Derivatives: A New Class of Non-Nucleoside Inhibitors Effective against a Broad Range of Drug-Resistant Strains.
J.Med.Chem., 48, 2005
|
|
1BQN
 
 | | TYR 188 LEU HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
1BQM
 
 | | HIV-1 RT/HBY 097 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Hsiou, Y, Das, K, Ding, J, Arnold, E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Tyr188Leu mutant and wild-type HIV-1 reverse transcriptase complexed with the non-nucleoside inhibitor HBY 097: inhibitor flexibility is a useful design feature for reducing drug resistance.
J.Mol.Biol., 284, 1998
|
|
4Q0B
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with gap-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-04-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4PWD
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with bulge-RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*U)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-19 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | Descriptor: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1K5M
 
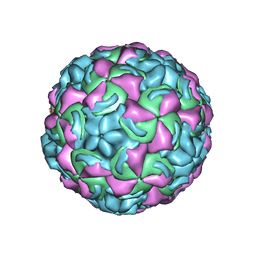 | | Crystal Structure of a Human Rhinovirus Type 14:Human Immunodeficiency Virus Type 1 V3 Loop Chimeric Virus MN-III-2 | | Descriptor: | CHIMERA OF HRV14 COAT PROTEIN VP2 (P1B) AND the V3 loop of HIV-1 gp120, COAT PROTEIN VP1 (P1D), COAT PROTEIN VP3 (P1C), ... | | Authors: | Ding, J, Smith, A.D, Geisler, S.C, Ma, X, Arnold, G.F, Arnold, E. | | Deposit date: | 2001-10-11 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Human Rhinovirus
that Displays Part of the HIV-1 V3 Loop and
Induces Neutralizing Antibodies against
HIV-1
Structure, 10, 2002
|
|
3BGR
 
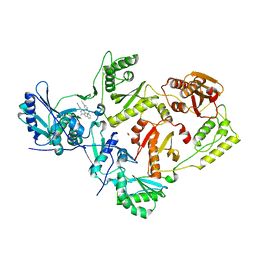 | | Crystal structure of K103N/Y181C mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (Rilpivirine), a non-nucleoside RT inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-11-27 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZE2
 
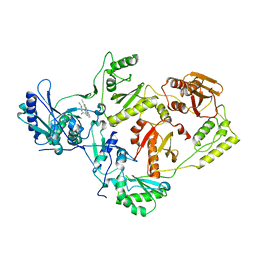 | | Crystal structure of L100I/K103N mutant HIV-1 reverse transcriptase (RT) in complex with TMC278 (rilpivirine), a non-nucleoside RT inhibitor | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-12-05 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2ZD1
 
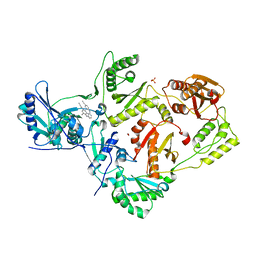 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with TMC278 (Rilpivirine), A Non-nucleoside RT Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, Reverse transcriptase/ribonuclease H, ... | | Authors: | Das, K, Bauman, J.D, Clark Jr, A.D, Shatkin, A.J, Arnold, E. | | Deposit date: | 2007-11-16 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: Strategic flexibility explains potency against resistance mutations.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
