7BHW
 
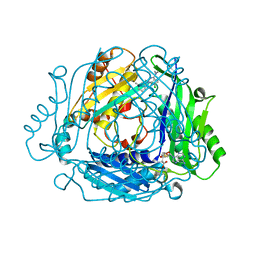 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 29) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-(3-methylphenyl)quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHR
 
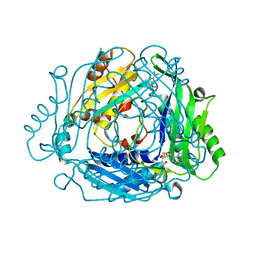 | | Crystal structure of MAT2a with triazinone fragment 1 bound in the allosteric site | | Descriptor: | 4-(dimethylamino)-6-ethoxy-1~{H}-1,3,5-triazin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHV
 
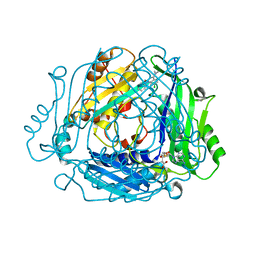 | | Crystal structure of MAT2a bound to allosteric inhibitor and in vivo tool compound 28 | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-phenyl-quinazolin-2-one, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHT
 
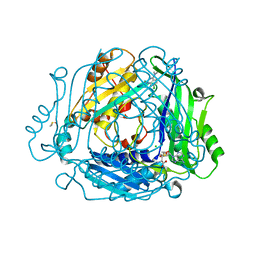 | | Crystal structure of MAT2a with quinazolinone fragment 5 bound in the allosteric site | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1~{H}-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.052 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7BHX
 
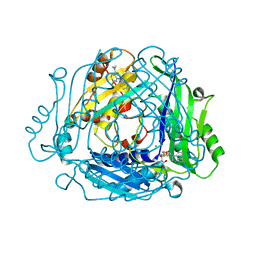 | | Crystal structure of MAT2a bound to allosteric inhibitor (compound 31) | | Descriptor: | 7-chloranyl-4-(dimethylamino)-1-pyridin-3-yl-quinazolin-2-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Schimpl, M, De Fusco, C, Borjesson, U, Cheung, T, Collie, I, Evans, L, Narasimhan, P, Stubbs, C, Vazquez-Chantada, M, Wagner, D.J, Grondine, M, Tentarelli, S, Underwood, E, Argyrou, A, Bagal, S, Chiarparin, E, Robb, G, Scott, J.S. | | Deposit date: | 2021-01-11 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment-Based Design of a Potent MAT2a Inhibitor and in Vivo Evaluation in an MTAP Null Xenograft Model.
J.Med.Chem., 64, 2021
|
|
7P1C
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with darobactin B | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Outer membrane protein assembly factor BamA, TRP-ASN-UX8-THR-LYS-ARG-PHE | | Authors: | Jakob, R.P, Modaresi, S.M, Hiller, S, Maier, T. | | Deposit date: | 2021-07-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mutasynthetic Production and Antimicrobial Characterization of Darobactin Analogs.
Microbiol Spectr, 9, 2021
|
|
5M2F
 
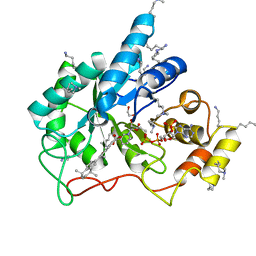 | | Crystal structure of human AKR1B10 complexed with NADP+ and the synthetic retinoid UVI2008 | | Descriptor: | 1,2-ETHANEDIOL, 3-bromo-4-[(1E)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)prop-1-en-1-yl]benzoic acid, Aldo-keto reductase family 1 member B10, ... | | Authors: | Ruiz, F.X, Cousido-Siah, A, Mitschler, A, Porte, S, Alvarez, S, Dominguez, M, Alvarez, R, de Lera, A.R, Pares, X, Farres, J, Podjarny, A. | | Deposit date: | 2016-10-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Structural basis for the inhibition of AKR1B10 by the C3 brominated TTNPB derivative UVI2008.
Chem. Biol. Interact., 276, 2017
|
|
7QFU
 
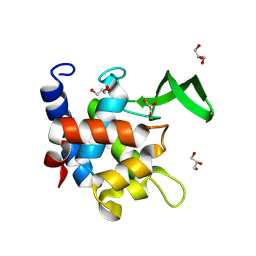 | | Crystal Structure of AtlA catalytic domain from Enterococcus feacalis | | Descriptor: | GLYCEROL, Peptidoglycan hydrolase | | Authors: | Zamboni, V, Barelier, S, Dixon, R, Galley, N, Ghanem, A, Cahuzac, H, Salamaga, B, Davis, P.J, Mesnage, S, Vincent, F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Molecular basis for substrate recognition and septum cleavage by AtlA, the major N-acetylglucosaminidase of Enterococcus faecalis.
J.Biol.Chem., 298, 2022
|
|
6ZMY
 
 | | Crystal structure of hemoglobin from turkey (Meleagiris gallopova) crystallized in monoclinic form at 1.66 Angstrom resolution | | Descriptor: | Hemoglobin beta chain, Hemoglobin subunit alpha-A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pandian, R, Shobana, N, Sundaresan, S.S, Thangaraj, V, Sayed, Y, Ponnuswamy, M.N. | | Deposit date: | 2020-07-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6XJ5
 
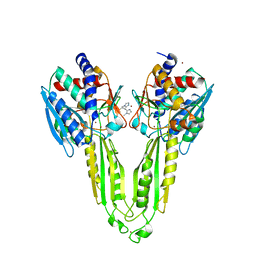 | |
6Y2K
 
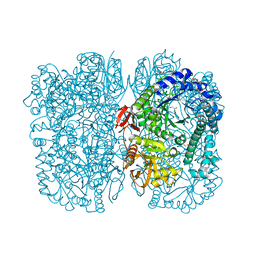 | | Crystal structure of beta-galactosidase from the psychrophilic Marinomonas ef1 | | Descriptor: | CHLORIDE ION, GLYCEROL, beta-galactosidase | | Authors: | Mangiagalli, M, Lapi, M, Maione, S, Orlando, M, Brocca, S, Pesce, A, Barbiroli, A, Pucciarelli, S, Camilloni, C, Lotti, M. | | Deposit date: | 2020-02-16 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement.
Febs J., 288, 2021
|
|
7AK9
 
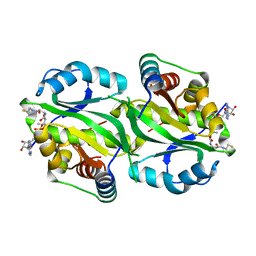 | | Structure of Salmonella TacT3 toxin bound to TacA3 antitoxin C-terminal peptide | | Descriptor: | ABC transporter, Acetyltransferase, DEPHOSPHO COENZYME A | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK7
 
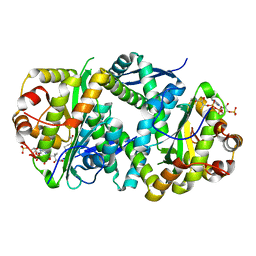 | | Structure of Salmonella TacT2 toxin bound to TacA2 antitoxin | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase, CHLORIDE ION, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Hare, S.A, Helaine, S. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
7AK8
 
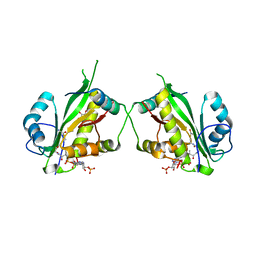 | | Structure of Salmonella TacT1 toxin bound to TacA1 antitoxin C-terminal peptide | | Descriptor: | ACETYL COENZYME *A, GCN5 family acetyltransferase, GLYCEROL, ... | | Authors: | Grabe, G.J, Morgan, R.M.L, Helaine, S, Hare, S.A. | | Deposit date: | 2020-09-30 | | Release date: | 2021-08-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Auxiliary interfaces support the evolution of specific toxin-antitoxin pairing.
Nat.Chem.Biol., 17, 2021
|
|
6QZJ
 
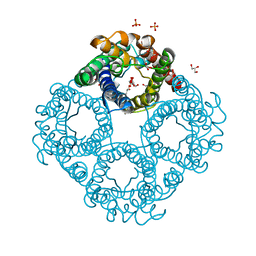 | | Crystal structure of human Aquaporin 7 at 2.2 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
6QZI
 
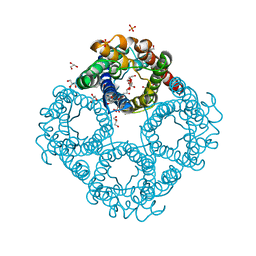 | | Crystal structure of human Aquaporin 7 at 1.9 A resolution | | Descriptor: | Aquaporin-7, GLYCEROL, PHOSPHATE ION | | Authors: | de Mare, S.W.-H, Venskutonyte, R, Eltschkner, S, Lindkvist-Petersson, K. | | Deposit date: | 2019-03-11 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Glycerol Efflux and Selectivity of Human Aquaporin 7.
Structure, 28, 2020
|
|
6ME7
 
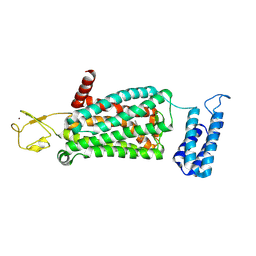 | | XFEL crystal structure of human melatonin receptor MT2 (H208A) in complex with 2-phenylmelatonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ... | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6ME6
 
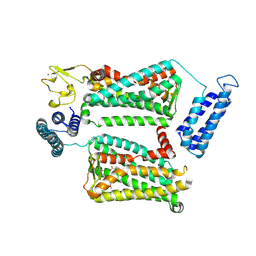 | | XFEL crystal structure of human melatonin receptor MT2 in complex with 2-phenylmelatonin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ... | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
6ME8
 
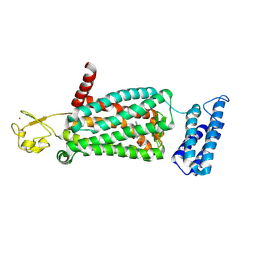 | | XFEL crystal structure of human melatonin receptor MT2 (N86D) in complex with 2-phenylmelatonin | | Descriptor: | N-[2-(5-methoxy-2-phenyl-1H-indol-3-yl)ethyl]acetamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
5O68
 
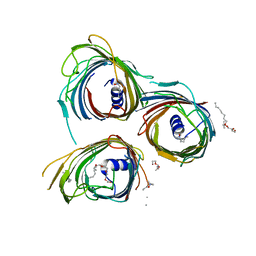 | | Crystal Structure of the Pseudomonas functional amyloid secretion protein FapF - R157A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
5O67
 
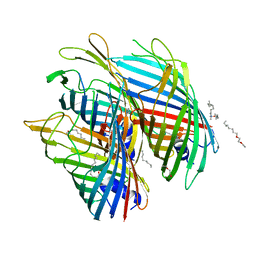 | | Crystal structure of the FapF polypeptide transporter - F103A mutant | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, FapF, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Rouse, S.L, Hare, S, Lambert, S, Morgan, R.M.L, Hawthorne, W.J, Berry, J, Matthews, S.J. | | Deposit date: | 2017-06-05 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A new class of hybrid secretion system is employed in Pseudomonas amyloid biogenesis.
Nat Commun, 8, 2017
|
|
7M6U
 
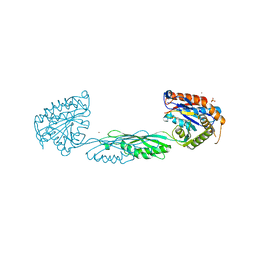 | |
6ME9
 
 | | XFEL crystal structure of human melatonin receptor MT2 in complex with ramelteon | | Descriptor: | N-{2-[(8S)-1,6,7,8-tetrahydro-2H-indeno[5,4-b]furan-8-yl]ethyl}propanamide, Soluble cytochrome b562,Melatonin receptor type 1B,Rubredoxin, ZINC ION | | Authors: | Johansson, L.C, Stauch, B, McCorvy, J, Han, G.W, Patel, N, Batyuk, A, Gati, C, Li, C, Grandner, J, Hao, S, Olsen, R.H.J, Tribo, A.R, Zaare, S, Zhu, L, Zatsepin, N.A, Weierstall, U, Liu, W, Roth, B.L, Katritch, V, Cherezov, V. | | Deposit date: | 2018-09-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | XFEL structures of the human MT2melatonin receptor reveal the basis of subtype selectivity.
Nature, 569, 2019
|
|
7S2H
 
 | | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation) | | Descriptor: | CITRATE ANION, Glucokinase 1, putative, ... | | Authors: | Kearns, S.P, Daneshian, L, Swartz, P.D, Carey, S.M, Chruszcz, M, D'Antonio, E.L. | | Deposit date: | 2021-09-03 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Trypanosoma cruzi glucokinase in the apo form (open conformation)
To Be Published
|
|
6Q3G
 
 | | Structure of native bacteriophage P68 | | Descriptor: | Arstotzka protein, Head fiber protein, Inner core protein, ... | | Authors: | Dominik, H, Karel, S, Fuzik, T, Plevka, P. | | Deposit date: | 2018-12-04 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and genome ejection mechanism ofStaphylococcus aureusphage P68.
Sci Adv, 5, 2019
|
|
