1KP8
 
 | |
1TYF
 
 | |
1F9S
 
 | | CRYSTAL STRUCTURE OF PLATELET FACTOR 4 MUTANT 2 | | Descriptor: | PLATELET FACTOR 4 | | Authors: | Yang, J, Doyle, M, Faulk, T, Visentin, G, Aster, R, Edwards, B. | | Deposit date: | 2000-07-11 | | Release date: | 2003-08-26 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structure Comparison of Two Platelet Factor 4 Mutants with the Wild-type Reveals the Epitopes for the Heparin-induced Thrombocytopenia Antibodies
To be Published
|
|
1UHV
 
 | | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase | | Descriptor: | 1,5-anhydro-2-deoxy-2-fluoro-D-xylitol, Beta-xylosidase | | Authors: | Yang, J.K, Yoon, H.J, Ahn, H.J, Il Lee, B, Pedelacq, J.D, Liong, E.C, Berendzen, J, Laivenieks, M, Vieille, C, Zeikus, G.J, Vocadlo, D.J, Withers, S.G, Suh, S.W. | | Deposit date: | 2003-07-11 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta-D-xylosidase from Thermoanaerobacterium saccharolyticum, a family 39 glycoside hydrolase.
J.Mol.Biol., 335, 2004
|
|
1UTA
 
 | | Solution structure of the C-terminal RNP domain from the divisome protein FtsN | | Descriptor: | CELL DIVISION PROTEIN FTSN | | Authors: | Yang, J.-C, van den Ent, F, Neuhaus, D, Brevier, J, Lowe, J. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Domain Architecture of the Divisome Protein Ftsn
Mol.Microbiol., 52, 2004
|
|
1RDQ
 
 | | Hydrolysis of ATP in the crystal of Y204A mutant of cAMP-dependent protein kinase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Ten Eyck, L.F, Xuong, N.H, Taylor, S.S. | | Deposit date: | 2003-11-05 | | Release date: | 2004-04-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal Structure of a cAMP-dependent Protein Kinase Mutant at 1.26A: New Insights into the Catalytic Mechanism.
J.Mol.Biol., 336, 2004
|
|
2WM9
 
 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GLYCEROL | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
1LRN
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1XX5
 
 | | Crystal Structure of Natrin from Naja atra snake venom | | Descriptor: | ETHANOL, Natrin 1 | | Authors: | Wang, J, Shen, B, Lou, X.H, Guo, M, Teng, M.K, Niu, L.W. | | Deposit date: | 2004-11-04 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from Naja atra venom that targets the BKCa channel
Biochemistry, 44, 2005
|
|
2AIR
 
 | |
2WMO
 
 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
1EQ1
 
 | |
2KAL
 
 | | NMR structure of fully methylated GATC site | | Descriptor: | 5'-D(*DCP*DGP*DCP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DCP*DGP*DC)-3', 5'-D(*DGP*DCP*DGP*DAP*DGP*(6MA)P*DTP*DCP*DTP*DGP*DCP*DG)-3' | | Authors: | Bang, J, Bae, S, Park, C, Lee, J, Choi, B. | | Deposit date: | 2008-11-09 | | Release date: | 2009-02-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural and dynamics study of DNA dodecamer duplexes that contain un-, hemi-, or fully methylated GATC sites.
J.Am.Chem.Soc., 130, 2008
|
|
1CS6
 
 | | N-TERMINAL FRAGMENT OF AXONIN-1 FROM CHICKEN | | Descriptor: | AXONIN-1, GLYCEROL | | Authors: | Freigang, J, Proba, K, Diederichs, K, Sonderegger, P, Welte, W. | | Deposit date: | 1999-08-17 | | Release date: | 2000-05-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion.
Cell(Cambridge,Mass.), 101, 2000
|
|
1Y5R
 
 | | The crystal structure of murine 11b-hydroxysteroid dehydrogenase complexed with corticosterone | | Descriptor: | CORTICOSTERONE, Corticosteroid 11-beta-dehydrogenase, isozyme 1, ... | | Authors: | Zhang, J, Osslund, T.D, Plant, M.H, Clogston, C.L, Nybo, R.E, Xiong, F, Delaney, J.M, Jordan, S. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Murine 11-Hydroxysteroid Dehydrogenase 1: An Important Therapeutic Target for Diabetes
Biochemistry, 44, 2005
|
|
1LRO
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP and Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1Q95
 
 | | Aspartate Transcarbamylase (ATCase) of Escherichia coli: A New Crystalline R State Bound to PALA, or to Product Analogues Phosphate and Citrate | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, N-(PHOSPHONACETYL)-L-ASPARTIC ACID, ... | | Authors: | Huang, J, Lipscomb, W.N. | | Deposit date: | 2003-08-22 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Aspartate Transcarbamylase (ATCase) of Escherichia coli: A New Crystalline R-State Bound to PALA, or to Product Analogues Citrate and Phosphate
Biochemistry, 43, 2004
|
|
1Y6V
 
 | | Structure of E. coli Alkaline Phosphatase in presence of cobalt at 1.60 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
1LRQ
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP, A5P and Cadmium | | Descriptor: | ARABINOSE-5-PHOSPHATE, CADMIUM ION, KDO-8-phosphate synthetase, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1QA9
 
 | | Structure of a Heterophilic Adhesion Complex Between the Human CD2 and CD58(LFA-3) Counter-Receptors | | Descriptor: | HUMAN CD2 PROTEIN, HUMAN CD58 PROTEIN | | Authors: | Wang, J.-H, Smolyar, A, Tan, K, Liu, J.-H, Kim, M, Sun, Z.J, Wagner, G, Reinherz, E.L. | | Deposit date: | 1999-04-13 | | Release date: | 1999-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors.
Cell(Cambridge,Mass.), 97, 1999
|
|
2XMQ
 
 | | Crystal structure of human NDRG2 protein provides insight into its role as a tumor suppressor | | Descriptor: | ACETATE ION, PROTEIN NDRG2 | | Authors: | Hwang, J, Kim, Y, Lee, H, Kim, M.H. | | Deposit date: | 2010-07-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Human Ndrg2 Protein Provides Insight Into its Role as a Tumor Suppressor.
J.Biol.Chem., 286, 2011
|
|
1JOO
 
 | | Averaged structure for unligated Staphylococcal nuclease-H124L | | Descriptor: | staphylococcal nuclease | | Authors: | Wang, J, Truckses, D.M, Abildgaard, F, Dzakula, Z, Zolnai, Z, Markley, J.L. | | Deposit date: | 2001-07-30 | | Release date: | 2001-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of staphylococcal nuclease from multidimensional, multinuclear NMR: nuclease-H124L and its ternary complex with Ca2+ and thymidine-3',5'-bisphosphate.
J.Biomol.NMR, 10, 1997
|
|
1Y5M
 
 | | The crystal structure of murine 11b-hydroxysteroid dehydrogenase: an important therapeutic target for diabetes | | Descriptor: | Corticosteroid 11-beta-dehydrogenase, isozyme 1, N-OCTANE, ... | | Authors: | Zhang, J, Osslund, T.D, Plant, M.H, Clogston, C.L, Nybo, R.E, Xiong, F, Delaney, J.M, Jordan, S.R. | | Deposit date: | 2004-12-02 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Murine 11-Hydroxysteroid Dehydrogenase 1: An Important Therapeutic Target for Diabetes
Biochemistry, 44, 2005
|
|
1JL4
 
 | | CRYSTAL STRUCTURE OF THE HUMAN CD4 N-TERMINAL TWO DOMAIN FRAGMENT COMPLEXED TO A CLASS II MHC MOLECULE | | Descriptor: | H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, A-K ALPHA CHAIN, A-K BETA CHAIN, ... | | Authors: | Wang, J.-H, Meijers, R, Reinherz, E.L. | | Deposit date: | 2001-07-15 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1Y7A
 
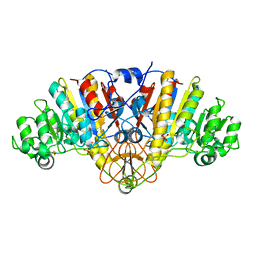 | | Structure of D153H/K328W E. coli alkaline phosphatase in presence of cobalt at 1.77 A resolution | | Descriptor: | Alkaline phosphatase, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Wang, J, Stieglitz, K, Kantrowitz, E.R. | | Deposit date: | 2004-12-08 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Metal Specificity Is Correlated with Two Crucial Active Site Residues in Escherichia coli Alkaline Phosphatase(,).
Biochemistry, 44, 2005
|
|
