6A8J
 
 | | Crystal structure of bacterial protein toxins | | Descriptor: | GLYCEROL, RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
4J2D
 
 | | RB69 DNA Polymerase L415K Ternary Complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*CP*GP*AP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Xia, S, Wang, J, Konigsberg, W.H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Role of L415 in the Replication of RB69 DNA Polymerase
To be Published
|
|
6L02
 
 | |
5XR4
 
 | | Crystal structure of RabA1a in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a, ... | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
2AVF
 
 | | Crystal Structure of C-terminal Desundecapeptide Nitrite Reductase from Achromobacter cycloclastes | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Li, H.T, Chang, T, Chang, W.C, Chen, C.J, Liu, M.Y, Gui, L.L, Zhang, J.P, An, X.M, Chang, W.R. | | Deposit date: | 2005-08-30 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of C-terminal desundecapeptide nitrite reductase from Achromobacter cycloclastes
Biochem.Biophys.Res.Commun., 338, 2005
|
|
5XX9
 
 | |
3P49
 
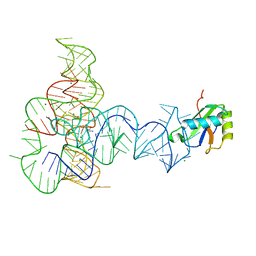 | | Crystal Structure of a Glycine Riboswitch from Fusobacterium nucleatum | | Descriptor: | GLYCINE, GLYCINE RIBOSWITCH, MAGNESIUM ION, ... | | Authors: | Butler, E.B, Wang, J, Xiong, Y, Strobel, S. | | Deposit date: | 2010-10-06 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structural basis of cooperative ligand binding by the glycine riboswitch.
Chem.Biol., 18, 2011
|
|
7E6R
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.6 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-23 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E6L
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.0 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78037143 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4ORB
 
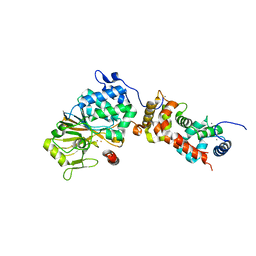 | | Crystal structure of mouse calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
7E6M
 
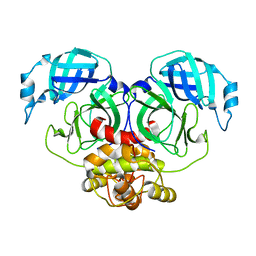 | | Crystal structure of Human coronavirus NL63 3C-like protease | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhong, F.L, Zhou, X.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83445024 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7E28
 
 | |
7E6N
 
 | | Crystal structure of HCoV-NL63 3C-like protease,pH5.2 | | Descriptor: | 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-02-22 | | Release date: | 2021-10-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8413 Å) | | Cite: | Crystal structures of human coronavirus NL63 main protease at different pH values
Acta Crystallogr.,Sect.F, 77, 2021
|
|
5XR7
 
 | | Crystal structure of RabA1a (Q72K) in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein RABA1a | | Authors: | Yun, J.S, Chang, J.H. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure and subcellular localization of RabA1a from Arabidopsis thaliana
To Be Published
|
|
4E02
 
 | | Crystal structure of branched-chain alpha-ketoacid dehydrogenase kinase/(S)-2-chloro-3-phenylpropanoic acid complex with AMPPNP | | Descriptor: | (S)-2-chloro-3-phenylpropanoic acid, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Tso, S.C, Chuang, J.L, Gui, W.J, Wynn, R.M, Li, J, Chuang, D.T. | | Deposit date: | 2012-03-02 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1547 Å) | | Cite: | Structures of branched-chain alpha-ketoacid dehydrogenase kinase-inhibitor complexes
To be Published
|
|
3LM5
 
 | | Crystal Structure of human Serine/Threonine Kinase 17B (STK17B) in complex with Quercetin | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Serine/threonine-protein kinase 17B | | Authors: | Ugochukwu, E, Soundararajan, M, Rellos, P, Fedorov, O, Phillips, C, Wang, J, Hapka, E, Filippakopoulos, P, Chaikuad, A, Pike, A.C.W, Carpenter, L, Vollmar, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Chemical Probe for Dark Kinase STK17B Derives Its Potency and High Selectivity through a Unique P-Loop Conformation.
J.Med.Chem., 63, 2020
|
|
6IJZ
 
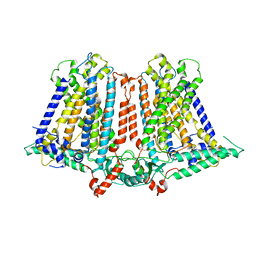 | | Structure of a plant cation channel | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Sun, L, Wang, J, Liu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2.
Nat Commun, 9, 2018
|
|
5GR8
 
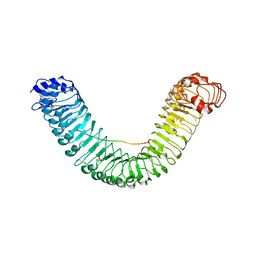 | | Crystal structure of PEPR1-AtPEP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Elicitor peptide 1, ... | | Authors: | Chai, J.J, Tang, J. | | Deposit date: | 2016-08-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1
Cell Res., 25, 2015
|
|
7D0F
 
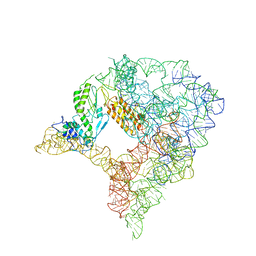 | | cryo-EM structure of a pre-catalytic group II intron RNP | | Descriptor: | Group II intron-encoded protein LtrA, RNA (738-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
6AYF
 
 | | TRPML3/ML-SA1 complex at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Mucolipin-3 | | Authors: | Zhou, X, Li, M, Su, D, Jia, Q, Li, H, Li, X, Yang, J. | | Deposit date: | 2017-09-08 | | Release date: | 2017-11-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM structures of the human endolysosomal TRPML3 channel in three distinct states.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7D0G
 
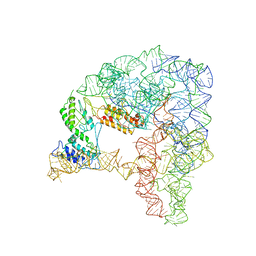 | | Cryo-EM structure of a pre-catalytic group II intron | | Descriptor: | Group II intron-encoded protein LtrA, RNA (714-MER) | | Authors: | Liu, N, Dong, X.L, Hu, C.X, Zeng, J.W, Wang, J.W, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2020-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Exon and protein positioning in a pre-catalytic group II intron RNP primed for splicing.
Nucleic Acids Res., 48, 2020
|
|
8SDU
 
 | | Structure of rat organic anion transporter 1 (OAT1) | | Descriptor: | Solute carrier family 22 member 6 | | Authors: | Dou, T, Jiang, J. | | Deposit date: | 2023-04-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | The substrate and inhibitor binding mechanism of polyspecific transporter OAT1 revealed by high-resolution cryo-EM.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SDZ
 
 | |
8SDY
 
 | |
4ORC
 
 | | Crystal structure of mammalian calcineurin | | Descriptor: | CALCIUM ION, Calcineurin subunit B type 1, FE (III) ION, ... | | Authors: | Ma, L, Li, S.J, Wang, J, Wu, J.W, Wang, Z.X. | | Deposit date: | 2014-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cooperative autoinhibition and multi-level activation mechanisms of calcineurin
To be Published
|
|
