4E3Q
 
 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
3NHG
 
 | | RB69 DNA Polymerase (S565G/Y567A) Ternary Complex with dTTP Opposite dG | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-14 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
3NE6
 
 | | RB69 DNA Polymerase (S565G/Y567A) Ternary Complex with dCTP Opposite dG | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-08 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
3NGI
 
 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dTTP Opposite dG | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-11 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.886 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
5YEH
 
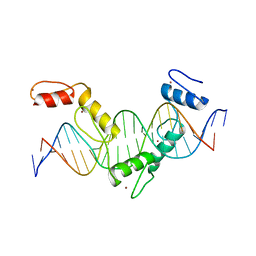 | | Crystal structure of CTCF ZFs4-8-eCBS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
7UFG
 
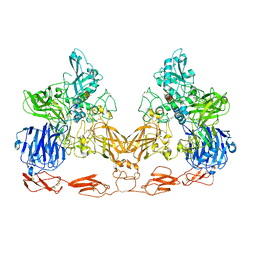 | | Cryo-EM structure of PAPP-A in complex with IGFBP5 | | Descriptor: | Insulin-like growth factor-binding protein 5, Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
2CEU
 
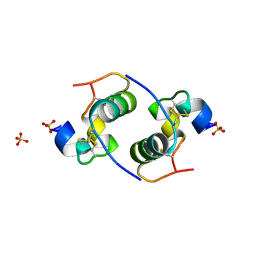 | | Despentapeptide insulin in acetic acid (pH 2) | | Descriptor: | INSULIN, SULFATE ION | | Authors: | Whittingham, J.L, Zhang, Y, Zakova, L, Dodson, E.J, Turkenburg, J.P, Brange, J, Dodson, G.G. | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | I222 Crystal Form of Despentapeptide (B26-B30) Insulin Provides New Insights Into the Properties of Monomeric Insulin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3NAE
 
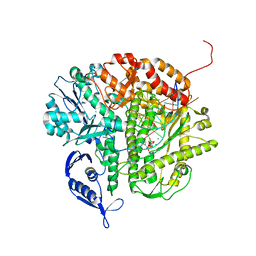 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dATP Opposite Guanidinohydantoin | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-01 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA Polymerase Allows dAMP and dGMP To Be Inserted opposite Guanidinohydantoin .
Biochemistry, 49, 2010
|
|
5LU5
 
 | | A quantum half-site enzyme | | Descriptor: | 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vivoli, M, Harmer, N.J, Pang, J. | | Deposit date: | 2016-09-08 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
6BVF
 
 | |
5VVO
 
 | |
5VVU
 
 | | Structural Investigations of the Substrate Specificity of Human O-GlcNAcase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-GlcNAcase, TAB1 peptide | | Authors: | Li, B, Jiang, J, Li, H, Hu, C.-W. | | Deposit date: | 2017-05-20 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase.
Nat Commun, 8, 2017
|
|
3LZJ
 
 | | RB69 DNA Polymerase (Y567A) ternary complex with dCTP Opposite 7,8-Dihydro-8-oxoguanine | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Beckman, J, Blaha, G, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-03-01 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution of Ala for Tyr567 in RB69 DNA polymerase allows dAMP to be inserted opposite 7,8-dihydro-8-oxoguanine .
Biochemistry, 49, 2010
|
|
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG7
 
 | |
8IG5
 
 | | Crystal structure of SARS main protease in complex with GC376 | | Descriptor: | 3C-like proteinase nsp5, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG6
 
 | | Crystal structure of MERS main protease in complex with GC376 | | Descriptor: | N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide, ORF1a | | Authors: | Lin, C, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
8IG8
 
 | |
2OQF
 
 | | Structure of a synthetic, non-natural analogue of RNase A: [N71K(Ade), D83A]RNase A | | Descriptor: | Ribonuclease pancreatic | | Authors: | Boerema, D.J, Tereshko, V.A, Zhang, J.L, He, C, Kent, S.B.H. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design, Synthesis, and Characterization of Non-natural RNase A Analogues with Enhanced Second-step Catalytic Activity
To be Published
|
|
5E8E
 
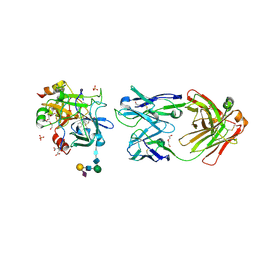 | | Crystal structure of thrombin bound to an exosite 1-specific IgA Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, ... | | Authors: | Baglin, T.P, Langdown, J, Frasson, R, Huntington, J.A. | | Deposit date: | 2015-10-14 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and characterization of an antibody directed against exosite I of thrombin.
J.Thromb.Haemost., 14, 2016
|
|
8IG9
 
 | |
2OUJ
 
 | |
8IGB
 
 | |
5VLY
 
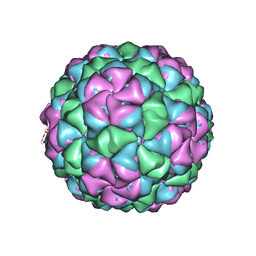 | | Asymmetric unit for the coat proteins of phage Qbeta | | Descriptor: | Capsid protein | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
