3IZM
 
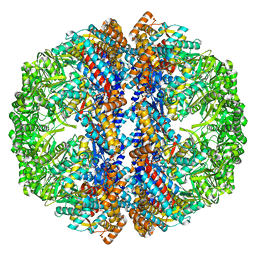 | | Mm-cpn wildtype with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (7.2 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZN
 
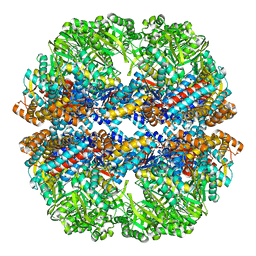 | | Mm-cpn deltalid with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-30 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.4 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZL
 
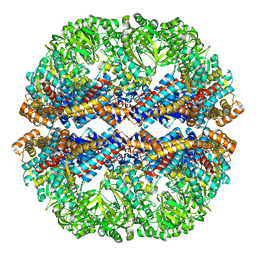 | | Mm-cpn rls deltalid with ATP and AlFx | | 分子名称: | Mm-cpn rls deltalid | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (6.2 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | 分子名称: | Chaperonin | | 著者 | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | 登録日 | 2010-10-29 | | 公開日 | 2011-02-02 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3J1T
 
 | | High affinity dynein microtubule binding domain - tubulin complex | | 分子名称: | Cytoplasmic dynein 1 heavy chain 1, seryl t-RNA synthetase chimera, Tubulin alpha-1B chain, ... | | 著者 | Redwine, W.B, Hernandez-Lopez, R, Zou, S, Huang, J, Reck-Peterson, S.L, Leschziner, A.E. | | 登録日 | 2012-06-25 | | 公開日 | 2012-09-26 | | 最終更新日 | 2024-02-21 | | 実験手法 | ELECTRON MICROSCOPY (9.7 Å) | | 主引用文献 | Structural basis for microtubule binding and release by dynein.
Science, 337, 2012
|
|
4D90
 
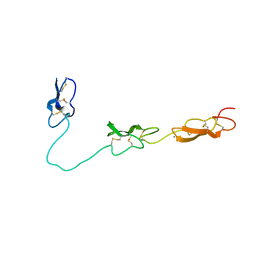 | | Crystal Structure of Del-1 EGF domains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | 著者 | Chen, Q, Schurpf, T, Springer, T, Wang, J. | | 登録日 | 2012-01-11 | | 公開日 | 2012-05-30 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | The RGD finger of Del-1 is a unique structural feature critical for integrin binding.
Faseb J., 26, 2012
|
|
1FBC
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | 分子名称: | 2,5-anhydro-1,6-di-O-phosphono-D-glucitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | 著者 | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | 登録日 | 1992-10-14 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
3WDU
 
 | | The complex structure of PtLic16A with cellobiose | | 分子名称: | Beta-1,3-1,4-glucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | 著者 | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | 登録日 | 2013-06-25 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
1FBG
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | 分子名称: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MANGANESE (II) ION | | 著者 | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | 登録日 | 1992-10-16 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
1P53
 
 | | The Crystal Structure of ICAM-1 D3-D5 fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1 | | 著者 | Yang, Y, Jun, C.D, Liu, J.H, Zhang, R, Jochimiak, A, Springer, T.A, Wang, J.H. | | 登録日 | 2003-04-24 | | 公開日 | 2004-05-04 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structural basis for dimerization of ICAM-1 on the cell surface.
Mol.Cell, 14, 2004
|
|
3WDT
 
 | | The apo-form structure of PtLic16A from Paecilomyces thermophila | | 分子名称: | Beta-1,3-1,4-glucanase, SULFATE ION | | 著者 | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | 登録日 | 2013-06-25 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
1FBF
 
 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | 分子名称: | 2,5-anhydro-1,6-di-O-phosphono-D-mannitol, FRUCTOSE 1,6-BISPHOSPHATASE, MAGNESIUM ION | | 著者 | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | 登録日 | 1992-10-16 | | 公開日 | 1993-10-31 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
3WP5
 
 | | The crystal structure of mutant CDBFV E109A from Neocallimastix patriciarum | | 分子名称: | CDBFV | | 著者 | Cheng, Y.S, Chen, C.C, Huang, C.H, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | 登録日 | 2014-01-09 | | 公開日 | 2014-03-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: insights into the molecular basis of a thermophilic enzyme.
J.Biol.Chem., 289, 2014
|
|
1FRP
 
 | | CRYSTAL STRUCTURE OF FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH FRUCTOSE-2,6-BISPHOSPHATE, AMP AND ZN2+ AT 2.0 ANGSTROMS RESOLUTION. ASPECTS OF SYNERGISM BETWEEN INHIBITORS | | 分子名称: | 2,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE, ... | | 著者 | Xue, Y, Huang, S, Liang, J.-Y, Zhang, Y, Lipscomb, W.N. | | 登録日 | 1994-08-26 | | 公開日 | 1994-11-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of fructose-1,6-bisphosphatase complexed with fructose 2,6-bisphosphate, AMP, and Zn2+ at 2.0-A resolution: aspects of synergism between inhibitors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
3WDY
 
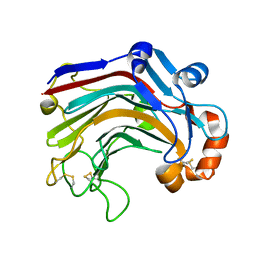 | | The complex structure of E113A with cellotetraose | | 分子名称: | Beta-1,3-1,4-glucanase, SULFATE ION, beta-D-glucopyranose, ... | | 著者 | Cheng, Y.S, Huang, C.H, Chen, C.C, Huang, T.Y, Ko, T.P, Huang, J.W, Wu, T.H, Liu, J.R, Guo, R.T. | | 登録日 | 2013-06-25 | | 公開日 | 2014-07-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Structural and mutagenetic analyses of a 1,3-1,4-beta-glucanase from Paecilomyces thermophila
Biochim.Biophys.Acta, 1844, 2014
|
|
2KZ1
 
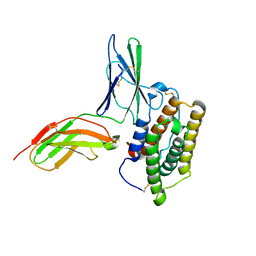 | | Inter-molecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric back-protonation and 2D NOESY | | 分子名称: | Interferon alpha-2, Soluble IFN alpha/beta receptor | | 著者 | Nudelman, I, Akabayov, S.R, Schnur, E, Biron, Z, Levy, R, Xu, Y, Yang, D, Anglister, J. | | 登録日 | 2010-06-10 | | 公開日 | 2010-06-23 | | 最終更新日 | 2021-08-18 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Intermolecular interactions in a 44 kDa interferon-receptor complex detected by asymmetric reverse-protonation and two-dimensional NOESY
Biochemistry, 49, 2010
|
|
3EXI
 
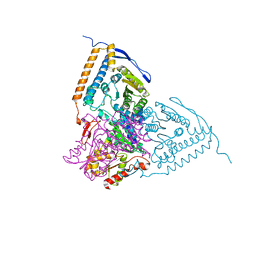 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex with the subunit-binding domain (SBD) of E2p, but SBD cannot be modeled into the electron density | | 分子名称: | CHLORIDE ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | 著者 | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | 登録日 | 2008-10-16 | | 公開日 | 2008-11-25 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
6DDT
 
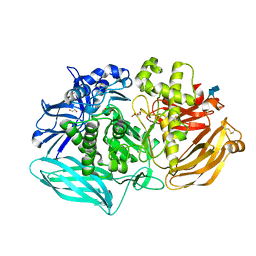 | | mouse beta-mannosidase (MANBA) | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gytz, H, Liang, J, Liang, Y, Gorelik, A, Illes, K, Nagar, B. | | 登録日 | 2018-05-10 | | 公開日 | 2019-01-09 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The structure of mammalian beta-mannosidase provides insight into beta-mannosidosis and nystagmus.
FEBS J., 286, 2019
|
|
4E9A
 
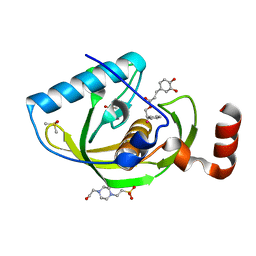 | | Structure of Peptide Deformylase form Helicobacter Pylori in complex with inhibitor | | 分子名称: | 2-phenylethyl (2E)-3-(3,4-dihydroxyphenyl)prop-2-enoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, COBALT (II) ION, ... | | 著者 | Cui, K, Zhu, L, Lu, W, Huang, J. | | 登録日 | 2012-03-20 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.662 Å) | | 主引用文献 | Identification of Novel Peptide Deformylase Inhibitors from Natural Products
To be Published
|
|
5Z3N
 
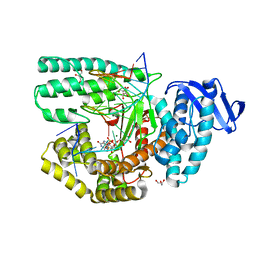 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base 5fC pair with dA | | 分子名称: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(5FC)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | 著者 | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | 登録日 | 2018-01-08 | | 公開日 | 2018-11-21 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
4EDT
 
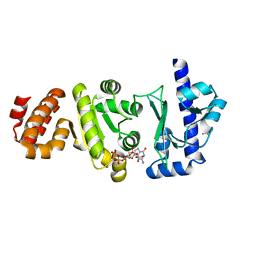 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to ppGpp and Manganese | | 分子名称: | BENZAMIDINE, DNA primase, GUANOSINE-5',3'-TETRAPHOSPHATE, ... | | 著者 | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | 登録日 | 2012-03-27 | | 公開日 | 2012-07-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.005 Å) | | 主引用文献 | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
4QAB
 
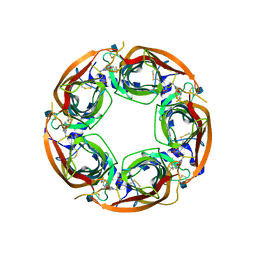 | | X-RAY STRUCTURE of ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 4-(MORPHOLIN-4-YL)-6-[4-(TRIFLUOROMETHYL)PHENYL]PYRIMIDIN-2-AMINE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(morpholin-4-yl)-6-[4-(trifluoromethyl)phenyl]pyrimidin-2-amine, Acetylcholine-binding protein, ... | | 著者 | Kaczanowska, K, Harel, M, Radic, Z, Changeux, J.-P, Finn, M.G, Taylor, P. | | 登録日 | 2014-05-03 | | 公開日 | 2014-07-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.984 Å) | | 主引用文献 | Structural basis for cooperative interactions of substituted 2-aminopyrimidines with the acetylcholine binding protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4EDR
 
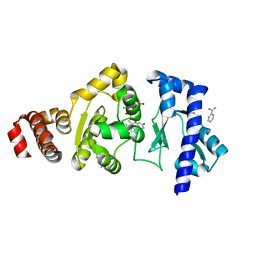 | | The structure of the S. aureus DnaG RNA Polymerase Domain bound to UTP and Manganese | | 分子名称: | BENZAMIDINE, DNA primase, MANGANESE (II) ION, ... | | 著者 | Rymer, R.U, Solorio, F.A, Chu, C, Corn, J.E, Wang, J.D, Berger, J.M. | | 登録日 | 2012-03-27 | | 公開日 | 2012-07-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Binding Mechanism of Metal-NTP Substrates and Stringent-Response Alarmones to Bacterial DnaG-Type Primases.
Structure, 20, 2012
|
|
7WKH
 
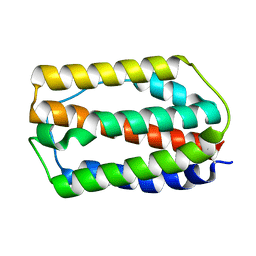 | | the grass carp IFNa | | 分子名称: | Interferon | | 著者 | Zou, J, WANG, J, Wang, Z. | | 登録日 | 2022-01-09 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Structural and Functional Analyses of Type I IFNa Shed Light Into Its Interaction With Multiple Receptors in Fish.
Front Immunol, 13, 2022
|
|
5ZQY
 
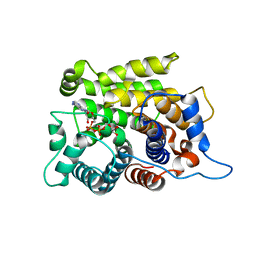 | | Crystal structure of a poly(ADP-ribose) glycohydrolase | | 分子名称: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | 著者 | Wang, M, Yuan, Z, Ma, Y, Wang, J, Liu, X. | | 登録日 | 2018-04-20 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.577 Å) | | 主引用文献 | Structure-function analyses reveal the mechanism of the ARH3-dependent hydrolysis of ADP-ribosylation.
J. Biol. Chem., 293, 2018
|
|
