1UW0
 
 | |
2B3V
 
 | | Spermine spermidine acetyltransferase in complex with acetylcoa, K26R mutant | | Descriptor: | ACETYL COENZYME *A, Diamine acetyltransferase 1 | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-21 | | Release date: | 2006-01-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2B4B
 
 | | SSAT+COA+BE-3-3-3, K26R mutant | | Descriptor: | COENZYME A, Diamine acetyltransferase 1, N-ETHYL-N-[3-(PROPYLAMINO)PROPYL]PROPANE-1,3-DIAMINE | | Authors: | Bewley, M.C, Graziano, V, Jiang, J.S, Matz, E, Studier, F.W, Pegg, A.P, Coleman, C.S, Flanagan, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-09-23 | | Release date: | 2006-01-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2VKM
 
 | | Crystal structure of GRL-8234 bound to BACE (Beta-secretase) | | Descriptor: | BETA-SECRETASE 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Hong, L, Tang, J, Ghosh, A.K. | | Deposit date: | 2007-12-04 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent Memapsin 2 (Beta-Secretase) Inhibitors: Design, Synthesis, Protein-Ligand X-Ray Structure, and in Vivo Evaluation.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1DU2
 
 | | SOLUTION STRUCTURE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Descriptor: | DNA POLYMERASE III | | Authors: | Keniry, M.A, Berthon, H.A, Yang, J.-Y, Miles, C.S, Dixon, N.E. | | Deposit date: | 2000-01-13 | | Release date: | 2000-05-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the theta subunit of DNA polymerase III from Escherichia coli.
Protein Sci., 9, 2000
|
|
3BQJ
 
 | | VA387 polypeptide | | Descriptor: | va387 polypeptide | | Authors: | Bu, W, Mamedova, A, Tan, M, Jiang, J, Hegde, R. | | Deposit date: | 2007-12-20 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
2XQU
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
1SJW
 
 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
1QU3
 
 | | INSIGHTS INTO EDITING FROM AN ILE-TRNA SYNTHETASE STRUCTURE WITH TRNA(ILE) AND MUPIROCIN | | Descriptor: | ISOLEUCYL-TRNA, ISOLEUCYL-TRNA SYNTHETASE, MUPIROCIN, ... | | Authors: | Silvian, L.F, Wang, J, Steitz, T.A. | | Deposit date: | 1999-07-06 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into editing from an ile-tRNA synthetase structure with tRNAile and mupirocin.
Science, 285, 1999
|
|
1JE9
 
 | | NMR SOLUTION STRUCTURE OF NT2 | | Descriptor: | SHORT NEUROTOXIN II | | Authors: | Cheng, Y, Wang, W, Wang, J. | | Deposit date: | 2001-06-16 | | Release date: | 2001-07-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
2AFJ
 
 | | SPRY domain-containing SOCS box protein 2 (SSB-2) | | Descriptor: | gene rich cluster, C9 gene | | Authors: | Masters, S.L, Yao, S, Willson, T.A, Zhang, J.G, Palmer, K.R, Smith, B.J, Babon, J.J, Nicola, N.A, Norton, R.S, Nicholson, S.E. | | Deposit date: | 2005-07-26 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The SPRY domain of SSB-2 adopts a novel fold that presents conserved Par-4-binding residues
Nat.Struct.Mol.Biol., 13, 2006
|
|
1ZXQ
 
 | | THE CRYSTAL STRUCTURE OF ICAM-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-2 | | Authors: | Casasnovas, J.M, Springer, T.A, Harrison, S.C, Wang, J.-H. | | Deposit date: | 1997-03-04 | | Release date: | 1997-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of ICAM-2 reveals a distinctive integrin recognition surface.
Nature, 387, 1997
|
|
1FZD
 
 | | STRUCTURE OF RECOMBINANT ALPHAEC DOMAIN FROM HUMAN FIBRINOGEN-420 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Spraggon, G, Applegate, D, Everse, S.J, Zhang, J.-Z, Veerapandian, L, Redman, C, Doolittle, R.F, Grieninger, G. | | Deposit date: | 1998-06-22 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a recombinant alphaEC domain from human fibrinogen-420.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
2XQT
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4, DICYCLOHEXYLUREA | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
1FXP
 
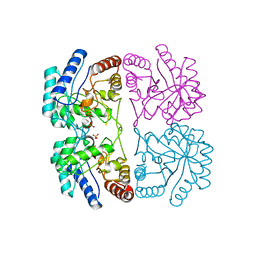 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH CADMIUM | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, CADMIUM ION, PHOSPHATE ION | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-26 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Substrate and metal complexes of 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus at 1.9-A resolution. Implications for the condensation mechanism.
J.Biol.Chem., 276, 2001
|
|
2KHS
 
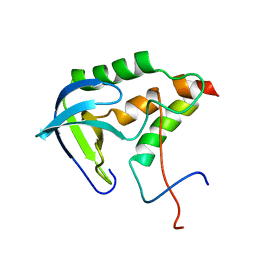 | | Solution structure of SNase121:SNase(111-143) complex | | Descriptor: | Nuclease, Thermonuclease | | Authors: | Geng, Y, Feng, Y, Xie, T, Shan, L, Wang, J. | | Deposit date: | 2009-04-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The native-like interactions between SNase121 and SNase(111-143) fragments induce the recovery of their native-like structures and the ability to degrade DNA.
Biochemistry, 48, 2009
|
|
2XQS
 
 | | Microscopic rotary mechanism of ion translocation in the Fo complex of ATP synthases | | Descriptor: | ATP SYNTHASE C CHAIN, CYMAL-4 | | Authors: | Pogoryelov, D, Krah, A, Langer, J, Yildiz, O, Faraldo-Gomez, J.D, Meier, T. | | Deposit date: | 2010-09-07 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microscopic Rotary Mechanism of Ion Translocation in the Fo Complex of ATP Synthases
Nat.Chem.Biol., 6, 2010
|
|
1ZA4
 
 | |
1ZJI
 
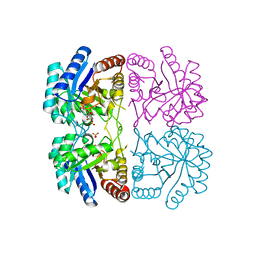 | | Aquifex aeolicus KDO8PS R106G mutant in complex with 2PGA and R5P | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop
Biochemistry, 44, 2005
|
|
1QRZ
 
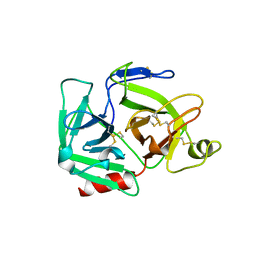 | | CATALYTIC DOMAIN OF PLASMINOGEN | | Descriptor: | PLASMINOGEN | | Authors: | Peisach, E, Wang, J, de los Santos, T, Reich, E, Ringe, D. | | Deposit date: | 1999-06-16 | | Release date: | 1999-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the proenzyme domain of plasminogen.
Biochemistry, 38, 1999
|
|
1FWN
 
 | | AQUIFEX AEOLICUS KDO8P SYNTHASE IN COMPLEX WITH PEP | | Descriptor: | 2-DEHYDRO-3-DEOXYPHOSPHOOCTONATE ALDOLASE, PHOSPHATE ION, PHOSPHOENOLPYRUVATE | | Authors: | Duewel, H.S, Radaev, S, Wang, J, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2000-09-23 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate and metal complexes of 3-deoxy-D-manno-octulosonate-8-phosphate synthase from Aquifex aeolicus at 1.9-A resolution. Implications for the condensation mechanism.
J.Biol.Chem., 276, 2001
|
|
2HR9
 
 | |
1ECL
 
 | |
1GZN
 
 | | Structure of PKB kinase domain | | Descriptor: | RAC-BETA SERINE/THREONINE PROTEIN KINASE | | Authors: | Barford, D, Yang, J, Hemmings, B.A. | | Deposit date: | 2002-05-24 | | Release date: | 2003-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Mechanism for the Regulation of Protein Kinase B/Akt by Hydrophobic Motif Phosphorylation
Mol.Cell, 9, 2002
|
|
1GZO
 
 | |
