5XGA
 
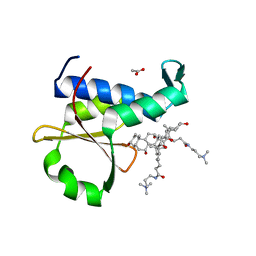 | | Crystal structure of the EnvZ periplasmic domain with CHAPS | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ACETIC ACID, Osmolarity sensor protein EnvZ | | Authors: | Hwang, E, Cheong, H.K, Jeon, Y.H, Cheong, C. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal structure of the EnvZ periplasmic domain with CHAPS.
FEBS Lett., 591, 2017
|
|
3BFP
 
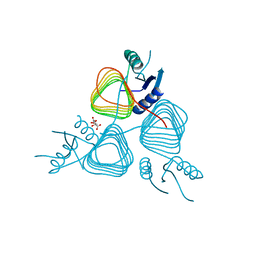 | | Crystal Structure of apo-PglD from Campylobacter jejuni | | Descriptor: | Acetyltransferase, CITRATE ANION | | Authors: | Rangarajan, E.S, Watson, D.C, Leclerc, S, Proteau, A, Cygler, M, Matte, A, Young, N.M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2007-11-22 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Active Site Residues of PglD, an N-Acetyltransferase from the Bacillosamine Synthetic Pathway Required for N-Glycan Synthesis in Campylobacter jejuni.
Biochemistry, 47, 2008
|
|
3SMZ
 
 | |
4W4Q
 
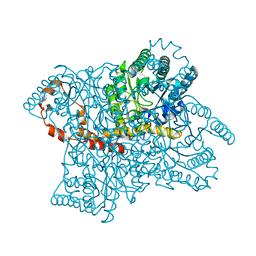 | | Glucose isomerase structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CALCIUM ION, Xylose isomerase | | Authors: | Nango, E, Tanaka, T, Sugahara, M, Suzuki, M, Iwata, S. | | Deposit date: | 2014-08-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat.Methods, 12, 2015
|
|
5VED
 
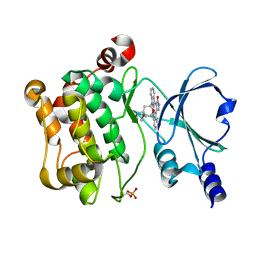 | |
8BWK
 
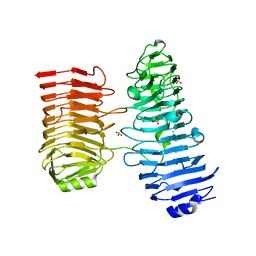 | |
7UXF
 
 | | Cryogenic electron microscopy 3D map of F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
7UTJ
 
 | | Cryogenic electron microscopy 3D map of F-actin bound by human dimeric alpha-catenin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Distinct inter-domain interactions of dimeric versus monomeric alpha-catenin link cell junctions to filaments.
Commun Biol, 6, 2023
|
|
3CM7
 
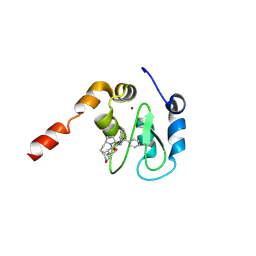 | | Crystal Structure of XIAP-BIR3 domain in complex with Smac-mimetic compuond, Smac005 | | Descriptor: | (3S,6S,7S,9aS)-6-{[(2S)-2-aminobutanoyl]amino}-N-(diphenylmethyl)-7-(hydroxymethyl)-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide, Baculoviral IAP repeat-containing protein 4, ZINC ION | | Authors: | Mastrangelo, E, Cossu, F, Milani, M. | | Deposit date: | 2008-03-21 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Targeting the X-linked inhibitor of apoptosis protein through 4-substituted azabicyclo[5.3.0]alkane smac mimetics. Structure, activity, and recognition principles.
J.Mol.Biol., 384, 2008
|
|
7UUW
 
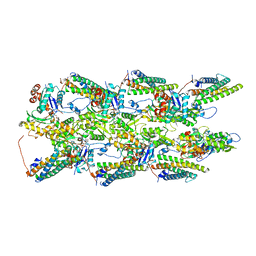 | | Cryogenic electron microscopy 3D map of F-actin bound by the Actin Binding Domain of alpha-catenin ortholog, HMP1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rangarajan, E.S, Smith, E.W, Izard, T. | | Deposit date: | 2022-04-29 | | Release date: | 2023-01-18 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | The nematode alpha-catenin ortholog, HMP1, has an extended alpha-helix when bound to actin filaments.
J.Biol.Chem., 299, 2022
|
|
5VEF
 
 | | PAK4 kinase domain in complex with fasudil | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, ACETATE ION, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhang, E.Y, Ha, B.H, Boggon, T.J. | | Deposit date: | 2017-04-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | PAK4 crystal structures suggest unusual kinase conformational movements.
Biochim. Biophys. Acta, 1866, 2018
|
|
5VEE
 
 | | PAK4 kinase domain in complex with FRAX486 | | Descriptor: | 6-(2,4-dichlorophenyl)-8-ethyl-2-{[3-fluoro-4-(piperazin-1-yl)phenyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhang, E.Y, Ha, B.H, Boggon, T.J. | | Deposit date: | 2017-04-04 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PAK4 crystal structures suggest unusual kinase conformational movements.
Biochim. Biophys. Acta, 1866, 2018
|
|
3LGE
 
 | | Crystal structure of rabbit muscle aldolase-SNX9 LC4 complex | | Descriptor: | Fructose-bisphosphate aldolase A, Sorting nexin-9 | | Authors: | Rangarajan, E.S, Park, H, Fortin, E, Sygusch, J, Izard, T. | | Deposit date: | 2010-01-20 | | Release date: | 2010-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of aldolase control of sorting nexin 9 function in endocytosis.
J.Biol.Chem., 285, 2010
|
|
4EC4
 
 | | XIAP-BIR3 in complex with a potent divalent Smac mimetic | | Descriptor: | (3S,6S,7S,9aS,3'S,6'S,7'S,9a'S)-N,N'-(benzene-1,4-diylbis{butane-4,1-diyl-1H-1,2,3-triazole-1,4-diyl[(S)-phenylmethanediyl]})bis[7-(hydroxymethyl)-6-{[(2S)-2-(methylamino)butanoyl]amino}-5-oxooctahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide], 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Baculoviral IAP repeat-containing protein 4, ... | | Authors: | Mastrangelo, E, Cossu, F, Bolognesi, M, Milani, M. | | Deposit date: | 2012-03-26 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural insight into inhibitor of apoptosis proteins recognition by a potent divalent smac-mimetic.
Plos One, 7, 2012
|
|
5GM0
 
 | | Tl-gal with lactose | | Descriptor: | beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, galectin | | Authors: | Hwang, E.Y. | | Deposit date: | 2016-07-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Carbohydrate Recognition and Anti-inflammatory Modulation by Gastrointestinal Nematode Parasite Toxascaris leonina Galectin
J. Biol. Chem., 291, 2016
|
|
6JZI
 
 | | Structure of hen egg-white lysozyme obtained from SFX experiments under atmospheric pressure | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Nango, E, Sugahara, M, Nakane, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
6JZH
 
 | | Structure of human A2A adenosine receptor in complex with ZM241385 obtained from SFX experiments under atmospheric pressure | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Nango, E, Shimamura, T, Nakane, T, Yamanaka, Y, Mori, C, Kimura, K.T, Fujiwara, T, Tanaka, T, Iwata, S. | | Deposit date: | 2019-05-02 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | High-viscosity sample-injection device for serial femtosecond crystallography at atmospheric pressure.
J.Appl.Crystallogr., 52, 2019
|
|
3HBN
 
 | | Crystal structure PseG-UDP complex from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, GLYCEROL, UDP-sugar hydrolase, ... | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
5B6V
 
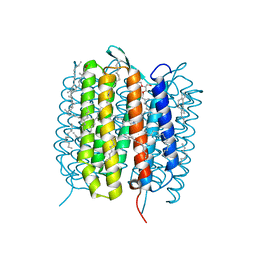 | | A three dimensional movie of structural changes in bacteriorhodopsin: resting state structure | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Nango, E, Royant, A, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
2PR3
 
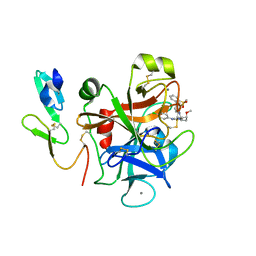 | | Factor XA inhibitor | | Descriptor: | (2R,4R)-N~1~-(4-CHLOROPHENYL)-N~2~-[3-FLUORO-2'-(METHYLSULFONYL)BIPHENYL-4-YL]-4-METHOXYPYRROLIDINE-1,2-DICARBOXAMIDE, CALCIUM ION, COAGULATION FACTOR X, ... | | Authors: | Zhang, E, Kohrt, J.T, Bigge, C.F, Finzel, B.C. | | Deposit date: | 2007-05-03 | | Release date: | 2007-08-14 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based drug design of pyrrolidine-1, 2-dicarboxamides as a novel series of orally bioavailable factor Xa inhibitors
Chem.Biol.Drug Des., 69, 2007
|
|
2PHB
 
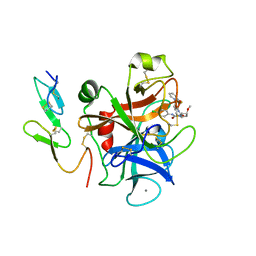 | | An Orally Efficacious Factor Xa Inhibitor | | Descriptor: | (2R,4R)-N~1~-(4-CHLOROPHENYL)-N~2~-[2-FLUORO-4-(2-OXOPYRIDIN-1(2H)-YL)PHENYL]-4-METHOXYPYRROLIDINE-1,2-DICARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Zhang, E, Kohrt, J.T, Bigge, C.F, Finzel, B.C. | | Deposit date: | 2007-04-10 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The discovery of (2R,4R)-N-(4-chlorophenyl)-N- (2-fluoro-4-(2-oxopyridin-1(2H)-yl)phenyl)-4-methoxypyrrolidine-1,2-dicarboxamide (PD 0348292), an orally efficacious factor Xa inhibitor
Chem.Biol.Drug Des., 70, 2007
|
|
3HBM
 
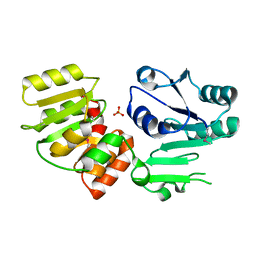 | | Crystal Structure of PseG from Campylobacter jejuni | | Descriptor: | SULFATE ION, UDP-sugar hydrolase | | Authors: | Rangarajan, E.S, Proteau, A, Cygler, M, Matte, A, Sulea, T, Schoenhofen, I.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-05-26 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of Campylobacter jejuni PseG: a udp-sugar hydrolase from the pseudaminic acid biosynthetic pathway.
J.Biol.Chem., 284, 2009
|
|
2OXT
 
 | | Crystal structure of Meaban virus nucleoside-2'-O-methyltransferase | | Descriptor: | NUCLEOSIDE-2'-O-METHYLTRANSFERASE, S-ADENOSYLMETHIONINE | | Authors: | Mastrangelo, E, Milani, M, Bollati, M, Bolognesi, M. | | Deposit date: | 2007-02-21 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural bases for substrate recognition and activity in Meaban virus nucleoside-2'-O-methyltransferase
Protein Sci., 16, 2007
|
|
4OH9
 
 | | Crystal Structure of the human MST2 SARAH homodimer | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Hwang, E, Cheong, H.-K, Ul Mushtaq, A, Kim, H.-Y, Yeo, K.J, Kim, E, Lee, W.C, Hwang, K.Y, Cheong, C, Jeon, Y.H. | | Deposit date: | 2014-01-17 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4YWU
 
 | | Structural insight into the substrate inhibition mechanism of NADP+-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes | | Descriptor: | 4-oxobutanoic acid, SULFATE ION, Succinic semialdehyde dehydrogenase | | Authors: | Jang, E.H, Park, S.A, Chi, Y.M, Lee, K.S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insight into the substrate inhibition mechanism of NADP(+)-dependent succinic semialdehyde dehydrogenase from Streptococcus pyogenes.
Biochem.Biophys.Res.Commun., 461, 2015
|
|
