2LV5
 
 | | NMR solution structure of PA1075 from Pseudomonas Aeruginosa | | Descriptor: | Uncharacterized protein | | Authors: | Andresen, C, Anandapadamanaban, M, Schneider, G, Schnell, R, Sunnerhagen, M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of PA1075, an essential protein in Pseudomonas Aeruginosa
To be Published
|
|
6RI7
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RH3
 
 | | Cryo-EM structure of E. coli RNA polymerase elongation complex bound to CTP substrate | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RI9
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in non-swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RIP
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex in swiveled state | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6RIN
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex bound to GreB transcription factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Abdelkareem, M, Saint-Andre, C, Takacs, M, Papai, G, Crucifix, C, Guo, X, Ortiz, J, Weixlbaumer, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of Transcription: RNA Polymerase Backtracking and Its Reactivation.
Mol.Cell, 75, 2019
|
|
6FVM
 
 | | Mutant DNA polymerase sliding clamp from Escherichia coli with bound P7 peptide | | Descriptor: | Beta sliding clamp, CALCIUM ION, GLYCEROL, ... | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
6FVL
 
 | | DNA polymerase sliding clamp from Escherichia coli with bound P7 peptide | | Descriptor: | Beta sliding clamp, GLYCEROL, P7 peptide, ... | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
6FVO
 
 | | Mutant DNA polymerase sliding clamp from Mycobacterium tuberculosis with bound P7 peptide | | Descriptor: | Beta sliding clamp, CALCIUM ION, P7 peptide | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
6FVN
 
 | | DNA polymerase sliding clamp from Mycobacterium tuberculosis with bound P7 peptide | | Descriptor: | Beta sliding clamp, P7 peptide | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.142 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
7AZ6
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 36 bound | | Descriptor: | ACETATE ION, Beta sliding clamp, CHLORIDE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ8
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 43 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZE
 
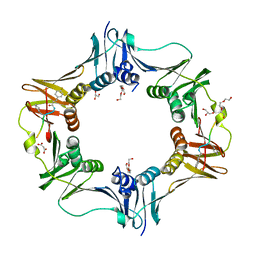 | | DNA polymerase sliding clamp from Escherichia coli with peptide 18 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, MALONATE ION, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ5
 
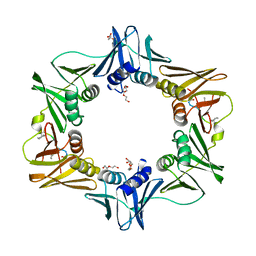 | | DNA polymerase sliding clamp from Escherichia coli with peptide 47 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, Peptide 47, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZG
 
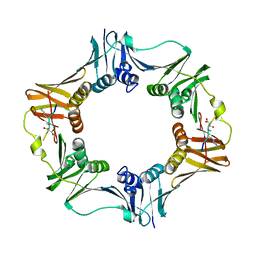 | | DNA polymerase sliding clamp from Escherichia coli with peptide 4 bound | | Descriptor: | Beta sliding clamp, Peptide 4 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZL
 
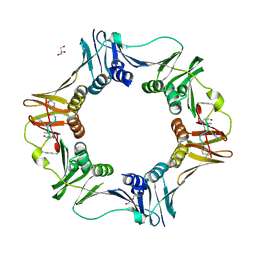 | | DNA polymerase sliding clamp from Escherichia coli with peptide 38 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZ7
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 37 bound | | Descriptor: | Beta sliding clamp, FORMIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZD
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 20 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZC
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 22 bound | | Descriptor: | Beta sliding clamp, GLYCEROL, Peptide 22 | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZF
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 8 bound | | Descriptor: | Beta sliding clamp, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
7AZK
 
 | | DNA polymerase sliding clamp from Escherichia coli with peptide 35 bound | | Descriptor: | Beta sliding clamp, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Monsarrat, C, Compain, G, Andre, C, Martiel, I, Engilberge, S, Olieric, V, Wolff, P, Brillet, K, Landolfo, M, Silva da Veiga, C, Wagner, J, Guichard, G, Burnouf, D.Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Iterative Structure-Based Optimization of Short Peptides Targeting the Bacterial Sliding Clamp.
J.Med.Chem., 64, 2021
|
|
1YKA
 
 | | Solution structure of Grx4, a monothiol glutaredoxin from E. coli. | | Descriptor: | monothiol glutaredoxin ydhD | | Authors: | Fladvad, M, Bellanda, M, Fernandes, A.P, Andresen, C, Mammi, S, Holmgren, A, Vlamis-Gardikas, A, Sunnerhagen, M. | | Deposit date: | 2005-01-17 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mapping of functionalities in the solution structure of reduced Grx4, a monothiol glutaredoxin from Escherichia coli.
J.Biol.Chem., 280, 2005
|
|
4AXT
 
 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME FROM AN AUTO HARVESTED CRYSTAL, Control Experiment | | Descriptor: | LYSOZYME C | | Authors: | Cipriani, F, Rower, M, Landret, C, Zander, U, Felisaz, F, Marquez, J.A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Direct: A New Method for Automated Crystal Harvesting Based on Laser-Induced Photoablation of Thin Films
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4B0D
 
 | | CRYSTAL STRUCTURE OF HEN EGG WHITE LYSOZYME FROM AN AUTO HARVESTED CRYSTAL | | Descriptor: | LYSOZYME C | | Authors: | Cipriani, F, Rower, M, Landret, C, Zander, U, Felisaz, F, Marquez, J.A. | | Deposit date: | 2012-07-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Direct: A New Method for Automated Crystal Harvesting Based on Laser-Induced Photoablation of Thin Films
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4B0A
 
 | | The high-resolution structure of yTBP-yTAF1 identifies conserved and competing interaction surfaces in transcriptional activation | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Anandapadamanaban, M, Andresen, C, Siponen, M, Kokubo, T, Ikura, M, Moche, M, Sunnerhagen, M. | | Deposit date: | 2012-06-29 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High-Resolution Structure of TBP with Taf1 Reveals Anchoring Patterns in Transcriptional Regulation
Nat.Struct.Mol.Biol., 20, 2013
|
|
