4ZV9
 
 | | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Filippova, E.V, Kiryukhina, O, Endres, M, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-18 | | Release date: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 2.00 Angstrom resolution crystal structure of an uncharacterized protein from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
5EZ4
 
 | | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
4ZWM
 
 | | 2.3 A resolution crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 | | Descriptor: | Ornithine aminotransferase, mitochondrial, putative, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Van Le, H, Silverman, R.B, McLeod, R, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-05-19 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 2.3 A resolution crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49
To Be Published
|
|
5EYU
 
 | | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
1TT4
 
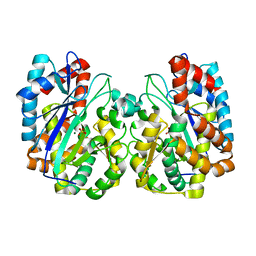 | | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium | | Descriptor: | MAGNESIUM ION, SULFATE ION, putative cytoplasmic protein | | Authors: | Miller, D.J, Shuvalova, L, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-21 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of NP459575, a predicted glutathione synthase from Salmonella typhimurium
To be Published
|
|
5CNP
 
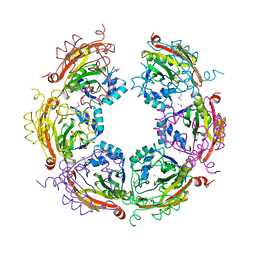 | | X-ray crystal structure of Spermidine n1-acetyltransferase from Vibrio cholerae. | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Osipiuk, J, VOLKART, L, MOY, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
5CJJ
 
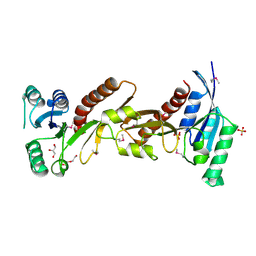 | | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-14 | | Release date: | 2015-07-29 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The crystal structure of phosphoribosylglycinamide formyltransferase from Campylobacter jejuni subsp. jejuni NCTC 11168
To Be Published
|
|
1SFS
 
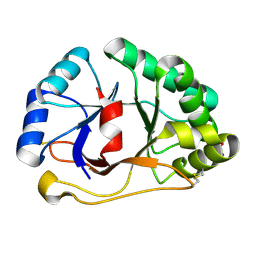 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
1RXQ
 
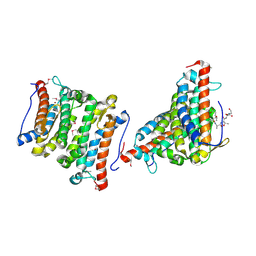 | | YfiT from Bacillus subtilis is a probable metal-dependent hydrolase with an unusual four-helix bundle topology | | Descriptor: | ALANINE, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Rajan, S.S, Yang, X, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-12-18 | | Release date: | 2004-07-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | YfiT from Bacillus subtilis Is a Probable Metal-Dependent Hydrolase with an Unusual Four-Helix Bundle Topology
Biochemistry, 43, 2004
|
|
5DGX
 
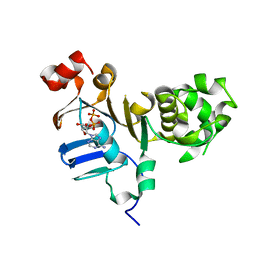 | | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipid A export ATP-binding/permease protein MsbA | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom resolution crystal structure of the ABC-ATPase domain (residues 357-609) of lipid A transport protein (msbA) from Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
5CRF
 
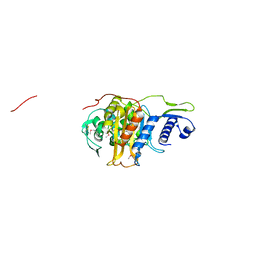 | | Structure of the penicillin-binding protein PonA1 from Mycobacterium Tuberculosis | | Descriptor: | PHOSPHATE ION, Penicillin-binding protein 1A | | Authors: | Filippova, E.V, Wawrzak, Z, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-22 | | Release date: | 2016-05-04 | | Last modified: | 2016-07-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5D6A
 
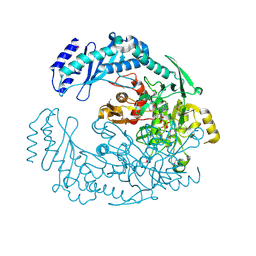 | | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP) | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Predicted ATPase of the ABC class, SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-11 | | Release date: | 2015-08-26 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom Crystal Structure of ABC transporter ATPase from Vibrio vulnificus in Complex with Adenylyl-imidodiphosphate (AMP-PNP)
To Be Published
|
|
5DJ9
 
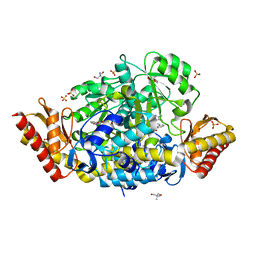 | | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with gabaculine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[O-PHOSPHONOPYRIDOXYL]--AMINO-BENZOIC ACID, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-01 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of the ornithine aminotransferase from Toxoplasma gondii ME49 in a complex with gabaculine
To Be Published
|
|
5CQE
 
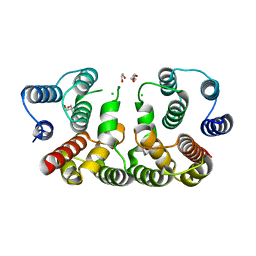 | | 2.1 Angstrom resolution crystal structure of matrix protein 1 (M1; residues 1-164) from Influenza A virus (A/Puerto Rico/8/34(H1N1)) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Flores, K, Dubrovska, I, Grimshaw, S, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of matrix protein 1 (M1; residues 1-164) from Influenza A virus (A/Puerto Rico/8/34(H1N1))
To Be Published
|
|
1TO9
 
 | |
5CXW
 
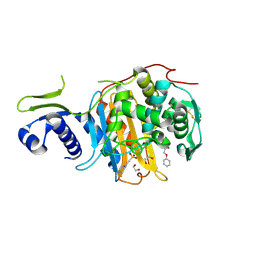 | | Structure of the PonA1 protein from Mycobacterium Tuberculosis in complex with penicillin V | | Descriptor: | (2R,4S)-5,5-dimethyl-2-{(1R)-2-oxo-1-[(phenoxyacetyl)amino]ethyl}-1,3-thiazolidine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Kiryukhina, O, Kieser, K, Endres, M, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2015-07-29 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance.
Febs J., 283, 2016
|
|
5BXI
 
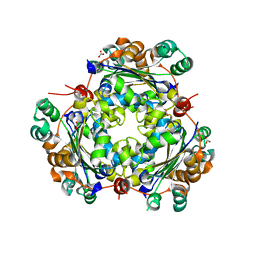 | | 1.7 Angstrom Resolution Crystal Structure of Putative Nucleoside Diphosphate Kinase from Toxoplasma gondii with Tyrosine of Tag Bound to Active Site | | Descriptor: | BICARBONATE ION, DI(HYDROXYETHYL)ETHER, Nucleoside diphosphate kinase | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-06-09 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
1SF9
 
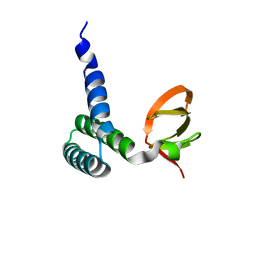 | | Crystal Structure of Bacillus subtilis YfhH Protein : Putative Transcriptional Regulator | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, yfhH hypothetical protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kim, D.E, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-19 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of Bacillus Subtilis YfhH hypothetical protein
To be Published
|
|
1U8X
 
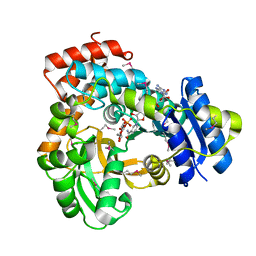 | | CRYSTAL STRUCTURE OF GLVA FROM BACILLUS SUBTILIS, A METAL-REQUIRING, NAD-DEPENDENT 6-PHOSPHO-ALPHA-GLUCOSIDASE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, MANGANESE (II) ION, Maltose-6'-phosphate glucosidase, ... | | Authors: | Rajan, S.S, Yang, X, Collart, F, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-09 | | Release date: | 2004-08-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Novel Catalytic Mechanism of Glycoside Hydrolysis Based on the Structure of an NAD(+)/Mn(2+)-Dependent Phospho-alpha-Glucosidase from Bacillus subtilis.
STRUCTURE, 12, 2004
|
|
1VRK
 
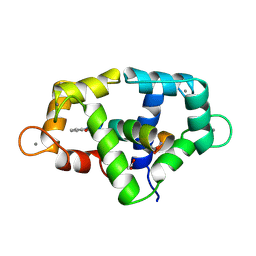 | |
1VR4
 
 | | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus | | Descriptor: | hypothetical protein APC22750 | | Authors: | Yang, X, Brunzelle, J.S, McNamara, L.K, Minasov, G, Shuvalova, L, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-01-31 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal Structure of MCSG TArget APC22750 from Bacillus cereus
To be Published
|
|
1EXC
 
 | | CRYSTAL STRUCTURE OF B. SUBTILIS MAF PROTEIN COMPLEXED WITH D-(UTP) | | Descriptor: | DEOXYURIDINE-5'-TRIPHOSPHATE, PROTEIN MAF, SODIUM ION | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EX2
 
 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS MAF PROTEIN | | Descriptor: | PHOSPHATE ION, PROTEIN MAF, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Minasov, G, Teplova, M, Stewart, G.C, Koonin, E.V, Anderson, W.F, Egli, M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-04-28 | | Release date: | 2000-06-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Functional implications from crystal structures of the conserved Bacillus subtilis protein Maf with and without dUTP.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5WOL
 
 | | Crystal structure of dihydrodipicolinate reductase DapB from Coxiella burnetii | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, 4-hydroxy-tetrahydrodipicolinate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyenko, O, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of dihydrodipicolinate reductase DapB from Coxiella burnetii
To Be Published
|
|
5WP0
 
 | | Crystal structure of NAD synthetase NadE from Vibrio fischeri | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Stogios, P.J, Evdokimova, E, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-03 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of NAD synthetase NadE from Vibrio fischeri
To Be Published
|
|
