3KQ4
 
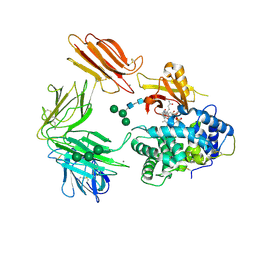 | | Structure of Intrinsic Factor-Cobalamin bound to its receptor Cubilin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Andersen, C.B.F, Madsen, M, Moestrup, S.K, Andersen, G.R. | | Deposit date: | 2009-11-17 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for receptor recognition of vitamin-B(12)-intrinsic factor complexes.
Nature, 464, 2010
|
|
8CI4
 
 | |
4WB3
 
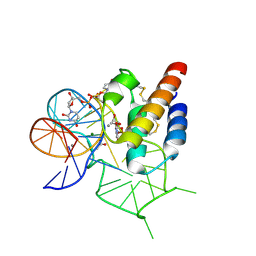 | | Crystal structure of the mirror-image L-RNA/L-DNA aptamer NOX-D20 in complex with mouse C5a-desArg complement anaphylatoxin | | Descriptor: | ACETATE ION, CALCIUM ION, Complement C5, ... | | Authors: | Yatime, L, Maasch, C, Hoehlig, K, Klussmann, S, Vater, A, Andersen, G.R. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer.
Nat Commun, 6, 2015
|
|
5NM0
 
 | | Nb36 Ser85Cys with Hg, crystal form 1 | | Descriptor: | MERCURY (II) ION, Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-06-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NML
 
 | | Nb36 Ser85Cys with Hg bound | | Descriptor: | 1,2-ETHANEDIOL, MERCURY (II) ION, Nanobody Nb36 Ser85Cys | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NLW
 
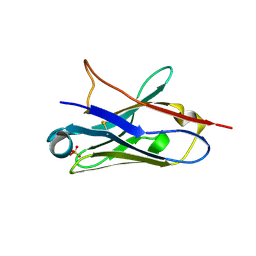 | | Structure of Nb36 crystal form 2 | | Descriptor: | SULFATE ION, nanobody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5NLU
 
 | | Structure of Nb36 crystal form 1 | | Descriptor: | SULFATE ION, single domain llama antibody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7NOZ
 
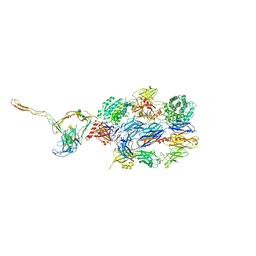 | | Structure of the nanobody stablized properdin bound alternative pathway proconvertase C3b:FB:FP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Lorenzen, J, Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2021-02-26 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structure determination of an unstable macromolecular complex enabled by nanobody-peptide bridging.
Protein Sci., 31, 2022
|
|
2IX8
 
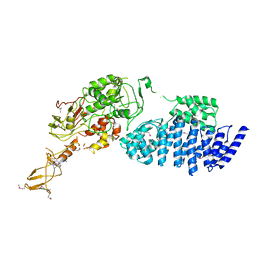 | | MODEL FOR EEF3 BOUND TO AN 80S RIBOSOME | | Descriptor: | ELONGATION FACTOR 3A | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-07 | | Release date: | 2007-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2IW3
 
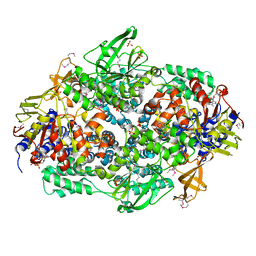 | | Elongation Factor 3 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ELONGATION FACTOR 3A, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-26 | | Release date: | 2006-08-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2IX3
 
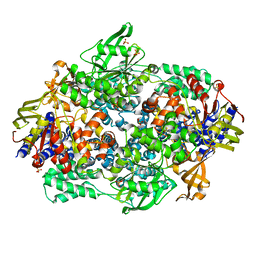 | | Structure of yeast Elongation Factor 3 | | Descriptor: | ELONGATION FACTOR 3, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-07-06 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2IWH
 
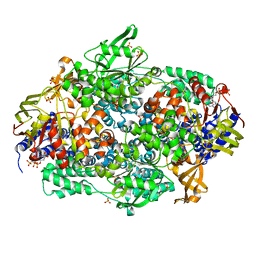 | | Structure of yeast Elongation Factor 3 in complex with ADPNP | | Descriptor: | ELONGATION FACTOR 3A, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION | | Authors: | Andersen, C.B.F, Becker, T, Blau, M, Anand, M, Halic, M, Balar, B, Mielke, T, Boesen, T, Pedersen, J.S, Spahn, C.M.T, Kinzy, T.G, Andersen, G.R, Beckmann, R. | | Deposit date: | 2006-06-30 | | Release date: | 2006-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Eef3 and the Mechanism of Transfer RNA Release from the E-Site.
Nature, 443, 2006
|
|
2HYI
 
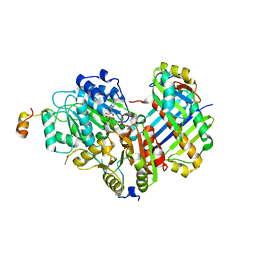 | | Structure of the human exon junction complex with a trapped DEAD-box helicase bound to RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*U)-3', MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Andersen, C.B.F, Le Hir, H, Andersen, G.R. | | Deposit date: | 2006-08-06 | | Release date: | 2006-08-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA.
Science, 313, 2006
|
|
7NP9
 
 | |
4WB2
 
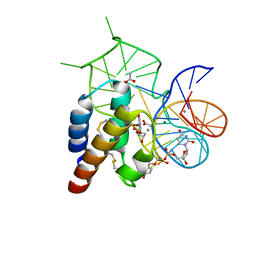 | | Crystal structure of the mirror-image L-RNA/L-DNA aptamer NOX-D20 in complex with mouse C5a complement anaphylatoxin | | Descriptor: | ACETATE ION, CALCIUM ION, Complement C5, ... | | Authors: | Yatime, L, Maasch, C, Hoehlig, K, Klussmann, S, Vater, A, Andersen, G.R. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer.
Nat Commun, 6, 2015
|
|
8OQ3
 
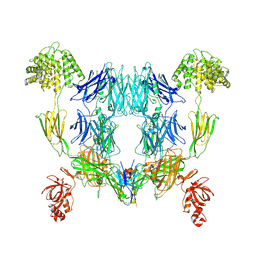 | | Structure of methylamine treated human complement C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gadeberg, T.A.F, Andersen, G.R. | | Deposit date: | 2023-04-11 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of methylamine treated human complement C3
To Be Published
|
|
4X7T
 
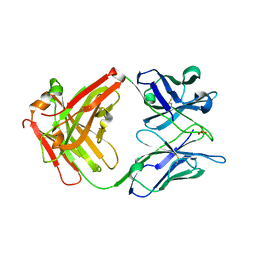 | |
4X7S
 
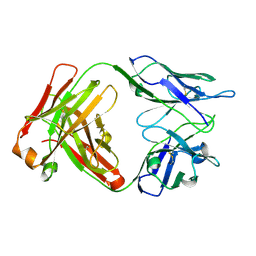 | |
4XW2
 
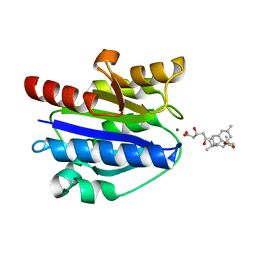 | | Structural basis for simvastatin competitive antagonism of complement receptor 3 | | Descriptor: | Integrin alpha-M, MAGNESIUM ION, Simvastatin acid | | Authors: | Bajic, G, Jensen, M.R, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2015-01-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Basis for Simvastatin Competitive Antagonism of Complement Receptor 3.
J.Biol.Chem., 291, 2016
|
|
7ZMK
 
 | |
4V4B
 
 | | Structure of the ribosomal 80S-eEF2-sordarin complex from yeast obtained by docking atomic models for RNA and protein components into a 11.7 A cryo-EM map. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0-A, 40S ribosomal protein S11, ... | | Authors: | Spahn, C.M, Gomez-Lorenzo, M.G, Grassucci, R.A, Jorgensen, R, Andersen, G.R, Beckmann, R, Penczek, P.A, Ballesta, J.P.G, Frank, J. | | Deposit date: | 2004-01-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation.
Embo J., 23, 2004
|
|
4LP4
 
 | |
2B7B
 
 | | Yeast guanine nucleotide exchange factor eEF1Balpha K205A mutant in complex with eEF1A and GDP | | Descriptor: | Elongation factor 1-alpha, GUANOSINE-5'-DIPHOSPHATE, elongation factor-1 beta | | Authors: | Pittman, Y.R, Valente, L, Jeppesen, M.G, Andersen, G.R, Patel, S, Kinzy, T.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha
J.Biol.Chem., 281, 2006
|
|
2B7C
 
 | | Yeast guanine nucleotide exchange factor eEF1Balpha K205A mutant in complex with eEF1A | | Descriptor: | Elongation factor 1-alpha, elongation factor-1 beta | | Authors: | Pittman, Y.R, Valente, L, Jeppesen, M.G, Andersen, G.R, Patel, S, Kinzy, T.G. | | Deposit date: | 2005-10-04 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mg2+ and a key lysine modulate exchange activity of eukaryotic translation elongation factor 1B alpha
J.Biol.Chem., 281, 2006
|
|
4M76
 
 | | Integrin I domain of complement receptor 3 in complex with C3d | | Descriptor: | Complement C3, Integrin alpha-M, NICKEL (II) ION | | Authors: | Bajic, G, Yatime, L, Vorup-Jensen, T, Andersen, G.R. | | Deposit date: | 2013-08-12 | | Release date: | 2013-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural insight on the recognition of surface-bound opsonins by the integrin I domain of complement receptor 3.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
