3U09
 
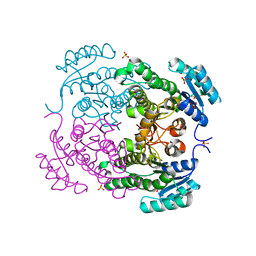 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG)(G92D) from Vibrio cholerae | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, SULFATE ION, UNKNOWN ATOM OR ION | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Grabowski, M, Fratczak, Z, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
1SFS
 
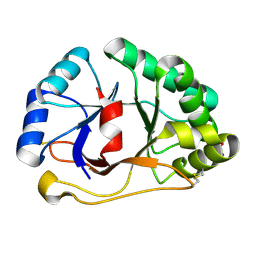 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
1TO9
 
 | |
3IGS
 
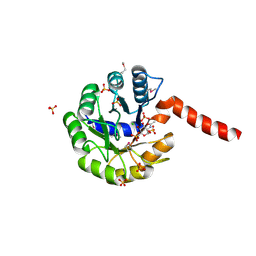 | | Structure of the Salmonella enterica N-acetylmannosamine-6-phosphate 2-epimerase | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, N-acetylmannosamine-6-phosphate 2-epimerase 2, ... | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Skarina, T, Papazisi, L, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-28 | | Release date: | 2009-08-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: |
|
|
3GOA
 
 | | Crystal structure of the Salmonella typhimurium FadA 3-ketoacyl-CoA thiolase | | Descriptor: | 3-ketoacyl-CoA thiolase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Anderson, S.M, Skarina, T, Onopriyenko, O, Wawrzak, Z, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: |
|
|
5UC7
 
 | | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex
To Be Published
|
|
5U08
 
 | | Crystal structure of an aminoglycoside acetyltransferase meta-AAC0020 from an uncultured soil metagenomic sample in complex with sisomicin | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, ACETATE ION, CALCIUM ION, ... | | Authors: | Xu, Z, Skarina, T, Wawrzak, Z, Stogios, P.J, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and Functional Survey of Environmental Aminoglycoside Acetyltransferases Reveals Functionality of Resistance Enzymes.
ACS Infect Dis, 3, 2017
|
|
5U1H
 
 | | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa | | Descriptor: | (2R,6S)-2-amino-6-(carboxyamino)-7-{[(1R)-1-carboxyethyl]amino}-7-oxoheptanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Watanabe, N, Stogios, P.J, Skarina, T, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-28 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the C-terminal peptidoglycan binding domain of OprF (PA1777) from Pseudomonas aeruginosa
To be published
|
|
5UB8
 
 | | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Likely rab family GTP-binding protein, ZINC ION | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-20 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of YPT31, a Rab family GTPase from Candida albicans, in complex with GDP and Zn(II)
To Be Published
|
|
5UE7
 
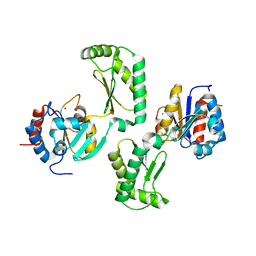 | | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of the phosphomannomutase PMM1 from Candida albicans, apoenzyme state
To Be Published
|
|
5UF8
 
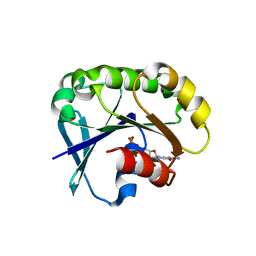 | | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Potential ADP-ribosylation factor | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-03 | | Release date: | 2017-01-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Crystal structure of the ARF family small GTPase ARF2 from Candida albicans in complex with GDP
To Be Published
|
|
5UYY
 
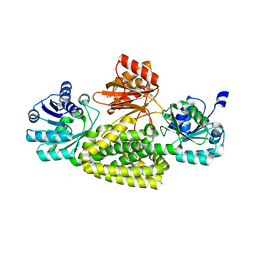 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with L-tyrosine | | Descriptor: | Prephenate dehydrogenase, TYROSINE | | Authors: | Shabalin, I.G, Hou, J, Cymborowski, M.T, Kwon, K, Christendat, D, Gritsunov, A.O, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
5V0V
 
 | | Crystal structure of Equine Serum Albumin complex with etodolac | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, Serum albumin, ... | | Authors: | Czub, M.P, Shabalin, I.G, Handing, K.B, Venkataramany, B.S, Steen, E.H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Albumin-Based Transport of Nonsteroidal Anti-Inflammatory Drugs in Mammalian Blood Plasma.
J.Med.Chem., 2020
|
|
5UM7
 
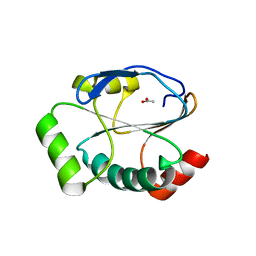 | | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii | | Descriptor: | ACETATE ION, Thioredoxin signature protein | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-26 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of the reduced state of the thiol-disulfide reductase SdbA from Streptococcus gordonii
To Be Published
|
|
5UTT
 
 | | SrtA sortase from Actinomyces oris | | Descriptor: | CHLORIDE ION, Sortase | | Authors: | Osipiuk, J, Ma, X, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cell-to-cell interaction requires optimal positioning of a pilus tip adhesin modulated by gram-positive transpeptidase enzymes.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5VKW
 
 | | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans | | Descriptor: | Adenylosuccinate lyase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-24 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of adenylosuccinate lyase ADE13 from Candida albicans
To Be Published
|
|
5VU3
 
 | | Crystal structure of the competence-damaged protein (CinA) superfamily protein ECL_02051 from Enterobacter cloacae | | Descriptor: | ACETATE ION, Competence damage-inducible protein A, TETRAETHYLENE GLYCOL | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Sandoval, J, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-18 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Crystal structure of the competence-damaged protein (CinA) superfamily protein ECL_02051 from Enterobacter cloacae
To Be Published
|
|
5VM2
 
 | | Crystal structure of ECK1772, an oxidoreductase/dehydrogenase of unknown specificity involved in membrane biogenesis from Escherichia coli | | Descriptor: | Alcohol dehydrogenase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-26 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Crystal structure of ECK1772, an oxidoreductase/dehydrogenase of unknown specificity involved in membrane biogenesis from Escherichia coli
To Be Published
|
|
4E1L
 
 | | Crystal structure of Acetoacetyl-CoA thiolase (thlA2) from Clostridium difficile | | Descriptor: | Acetoacetyl-CoA thiolase 2, IODIDE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: |
|
|
5VQB
 
 | | Crystal structure of rifampin monooxygenase from Streptomyces venezuelae, complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Cox, G, Kelso, J, Stogios, P.J, Savchenko, A, Anderson, W.F, Wright, G.D, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.391 Å) | | Cite: | Rox, a Rifamycin Resistance Enzyme with an Unprecedented Mechanism of Action.
Cell Chem Biol, 25, 2018
|
|
5VGC
 
 | | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Borek, D, Valleau, D, Skarina, T, Jobin, M.C, Wawrzak, Z, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
5U8J
 
 | | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae | | Descriptor: | CHLORIDE ION, UPF0502 protein BFJ73_07745 | | Authors: | Stogios, P.J, Skarina, T, Sandoval, J, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of a protein of unknown function ECL_02571 involved in membrane biogenesis from Enterobacter cloacae
To Be Published
|
|
5VYV
 
 | | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli | | Descriptor: | DI(HYDROXYETHYL)ETHER, UPF0502 protein YceH | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-26 | | Release date: | 2017-06-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of a protein of unknown function YceH/ECK1052 involved in membrane biogenesis from Escherichia coli
To Be Published
|
|
5VGM
 
 | | Crystal structure of dihydroorotase pyrC from Vibrio cholerae in complex with zinc at 1.95 A resolution. | | Descriptor: | ACETATE ION, CHLORIDE ION, Dihydroorotase, ... | | Authors: | Lipowska, J, Shabalin, I.G, Miks, C.D, Winsor, J, Cooper, D.R, Shuvalova, L, Kwon, K, Lewinski, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-11 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pyrimidine biosynthesis in pathogens - Structures and analysis of dihydroorotases from Yersinia pestis and Vibrio cholerae.
Int.J.Biol.Macromol., 136, 2019
|
|
5UXA
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
