6GL7
 
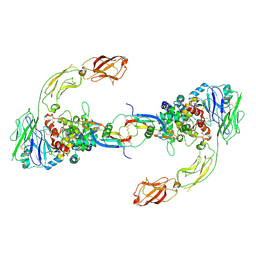 | | Neurturin-GFRa2-RET extracellular complex | | Descriptor: | GDNF family receptor alpha-2, Neurturin, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Bigalke, J.M, Aibara, S, Sandmark, J, Amunts, A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-08-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the activated RET signaling complex reveals the importance of its cysteine-rich domain.
Sci Adv, 5, 2019
|
|
3J7Y
 
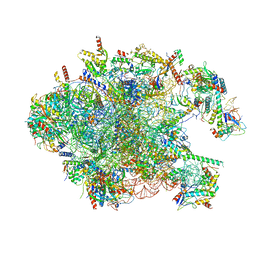 | | Structure of the large ribosomal subunit from human mitochondria | | Descriptor: | 16S rRNA, ADENOSINE MONOPHOSPHATE, CRIF1, ... | | Authors: | Brown, A, Amunts, A, Bai, X.C, Sugimoto, Y, Edwards, P.C, Murshudov, G, Scheres, S.H.W, Ramakrishnan, V. | | Deposit date: | 2014-08-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the large ribosomal subunit from human mitochondria.
Science, 346, 2014
|
|
8B6F
 
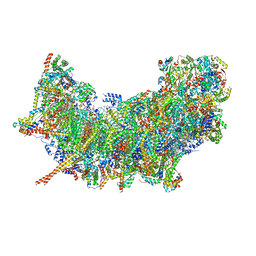 | | Cryo-EM structure of NADH:ubiquinone oxidoreductase (complex-I) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6H
 
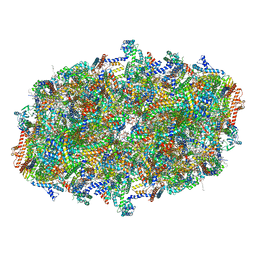 | | Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6G
 
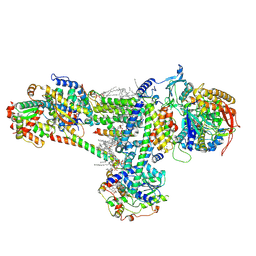 | | Cryo-EM structure of succinate dehydrogenase complex (complex-II) in respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CALCIUM ION, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8B6J
 
 | | Cryo-EM structure of cytochrome bc1 complex (complex-III) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apocytochrome b, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
5NGM
 
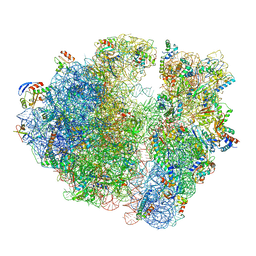 | | 2.9S structure of the 70S ribosome composing the S. aureus 100S complex | | Descriptor: | 16S ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Aibara, S, Zimmerman, E, Bashan, A, Amunts, A, Yonath, A. | | Deposit date: | 2017-03-18 | | Release date: | 2017-10-04 | | Last modified: | 2018-11-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The cryo-EM structure of hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.
Nat Commun, 8, 2017
|
|
5MRF
 
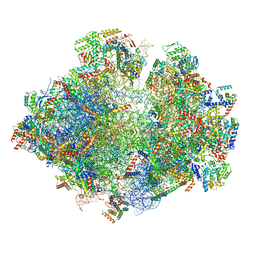 | | Structure of the yeast mitochondrial ribosome - Class C | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.97 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
5MRE
 
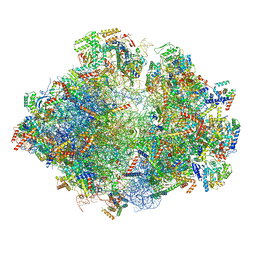 | | Structure of the yeast mitochondrial ribosome - Class B | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
5MRC
 
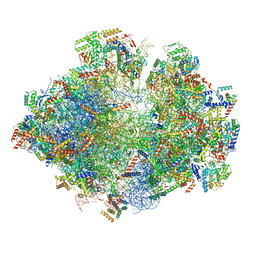 | | Structure of the yeast mitochondrial ribosome - Class A | | Descriptor: | 15S ribosomal RNA, 21S ribosomal RNA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Desai, N, Brown, A, Amunts, A, Ramakrishnan, V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-02-15 | | Last modified: | 2018-02-07 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structure of the yeast mitochondrial ribosome.
Science, 355, 2017
|
|
8BQS
 
 | | Cryo-EM structure of the I-II-III2-IV2 respiratory supercomplex from Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2 iron, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Baradaran, R, Amunts, A. | | Deposit date: | 2022-11-21 | | Release date: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mitochondrial membrane bending by I-II-III2-IV2 supercomplex
To Be Published
|
|
8BCW
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
8BCV
 
 | | Photosystem I assembly intermediate of Avena sativa | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A, Nelson, N. | | Deposit date: | 2022-10-17 | | Release date: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Photosystem I assembly intermediate of Avena sativa
To Be Published
|
|
8ANY
 
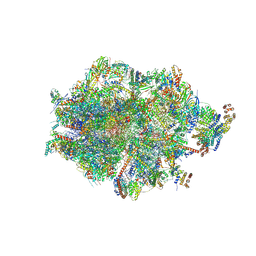 | | Human mitochondrial ribosome in complex with LRPPRC, SLIRP, A-site, P-site, E-site tRNAs and mRNA | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2022-08-06 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of LRPPRC-SLIRP-dependent translation by the mitoribosome.
Nat.Struct.Mol.Biol., 2024
|
|
6I9R
 
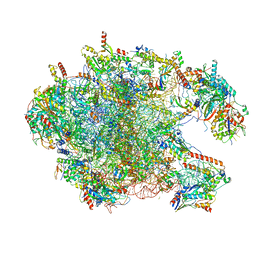 | | Large subunit of the human mitochondrial ribosome in complex with Virginiamycin M and Quinupristin | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Modelska, A, Aibara, S, Amunts, A. | | Deposit date: | 2018-11-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Inhibition of mitochondrial translation suppresses glioblastoma stem cell growth.
Cell Rep, 35, 2021
|
|
8APO
 
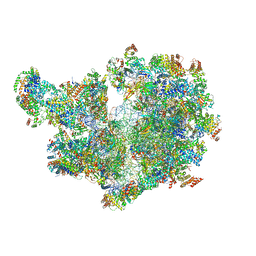 | | Structure of the mitochondrial ribosome from Polytomella magna with tRNAs bound to the A and P sites | | Descriptor: | A-site tRNA anticodon loop, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tobiasson, V, Berzina, I, Amunts, A. | | Deposit date: | 2022-08-10 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a mitochondrial ribosome with fragmented rRNA in complex with membrane-targeting elements.
Nat Commun, 13, 2022
|
|
7QI6
 
 | | Human mitochondrial ribosome in complex with mRNA, A/P- and P/E-tRNAs at 2.98 A resolution | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk.
Nat Commun, 15, 2024
|
|
7QI5
 
 | | Human mitochondrial ribosome in complex with mRNA, A/A-, P/P- and E/E-tRNAs at 2.63 A resolution | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-14 | | Release date: | 2023-07-05 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk.
Nat Commun, 15, 2024
|
|
7QI4
 
 | | Human mitochondrial ribosome at 2.2 A resolution (bound to partly built tRNAs and mRNA) | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-14 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Mitoribosome structure with cofactors and modifications reveals mechanism of ligand binding and interactions with L1 stalk.
Nat Commun, 15, 2024
|
|
6ZM5
 
 | | Human mitochondrial ribosome in complex with OXA1L, mRNA, A/A tRNA, P/P tRNA and nascent polypeptide | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Andrell, J, Amunts, A. | | Deposit date: | 2020-07-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Mechanism of membrane-tethered mitochondrial protein synthesis.
Science, 371, 2021
|
|
7AOI
 
 | | Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Tobiasson, V, Gahura, O, Aibara, S, Baradaran, R, Zikova, A, Amunts, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit.
Embo J., 40, 2021
|
|
6ZSE
 
 | | Human mitochondrial ribosome in complex with mRNA, A/P-tRNA and P/E-tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
6ZSB
 
 | | Human mitochondrial ribosome in complex with mRNA and P-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
6ZSG
 
 | | Human mitochondrial ribosome in complex with mRNA, A-site tRNA, P-site tRNA and E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
6ZSA
 
 | | Human mitochondrial ribosome bound to mRNA, A-site tRNA and P-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
