4HL9
 
 | | Crystal structure of antibiotic biosynthesis monooxygenase | | Descriptor: | Antibiotic biosynthesis monooxygenase | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of antibiotic biosynthesis monooxygenase
To be Published
|
|
3TPC
 
 | | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021 | | Descriptor: | Short chain alcohol dehydrogenase-related dehydrogenase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-07 | | Release date: | 2011-09-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal structure of a hypothtical protein SMa1452 from Sinorhizobium meliloti 1021
To be Published
|
|
4M1V
 
 | | Crystal structure of the ancestral soluble variant of the Human Phosphate Binding Protein (HPBP) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Gonzalez, D, Hiblot, J, Darbinian, N, Miller, J.S, Gotthard, G, Amini, S, Chabriere, E, Elias, M. | | Deposit date: | 2013-08-04 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Ancestral mutations as a tool for solubilizing proteins: The case of a hydrophobic phosphate-binding protein.
FEBS Open Bio, 4, 2014
|
|
3U5R
 
 | |
3U9L
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-19 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
3TXV
 
 | | Crystal structure of a probable tagatose 6 phosphate kinase from Sinorhizobium meliloti 1021 | | Descriptor: | Probable tagatose 6-phosphate kinase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a probable tagatose 6 phosphate kinase from Sinorhizobium meliloti 1021
To be Published
|
|
1SG9
 
 | | Crystal structure of Thermotoga maritima protein HEMK, an N5-glutamine methyltransferase | | Descriptor: | GLUTAMINE, S-ADENOSYLMETHIONINE, hemK protein | | Authors: | Agarwal, R, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-02-23 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel mode of dimerization via formation of a glutamate anhydride crosslink in a protein crystal structure.
Proteins, 71, 2008
|
|
1SGM
 
 | |
1T3A
 
 | | Crystal structure of Clostridium botulinum neurotoxin type E catalytic domain | | Descriptor: | CHLORIDE ION, ZINC ION, neurotoxin type E | | Authors: | Agarwal, R, Eswaramoorthy, S, Kumaran, D, Binz, T, Swaminathan, S. | | Deposit date: | 2004-04-26 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of botulinum neurotoxin type E catalytic domain and its mutant Glu212-->Gln reveals the pivotal role of the Glu212 carboxylate in the catalytic pathway
Biochemistry, 43, 2004
|
|
2HAF
 
 | | Crystal structure of a putative translation repressor from Vibrio cholerae | | Descriptor: | Putative translation repressor | | Authors: | Sugadev, R, Seetharaman, J, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of a putative translation repressor from Vibrio cholerae
To be Published
|
|
4J2U
 
 | | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1 | | Descriptor: | Enoyl-CoA hydratase | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Al Obaidi, N, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-02-05 | | Release date: | 2013-02-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an enoyl-CoA hydratase from Rhodobacter sphaeroides 2.4.1
To be Published
|
|
3T8L
 
 | | Crystal Structure of adenine deaminase with Mn/Fe | | Descriptor: | Adenine deaminase 2, UNKNOWN ATOM OR ION | | Authors: | Bagaria, A, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The catalase activity of diiron adenine deaminase.
Protein Sci., 20, 2011
|
|
3TFO
 
 | | Crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein) reductase from Sinorhizobium meliloti | | Descriptor: | HEXANE-1,6-DIOL, SULFATE ION, putative 3-oxoacyl-(acyl-carrier-protein) reductase | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-16 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-(acyl-carrier-protein) reductase from Sinorhizobium meliloti
To be Published
|
|
4JX9
 
 | | Crystal structure of the complex of peptidyl t-RNA hydrolase from Acinetobacter baumannii with uridine at 1.4A resolution | | Descriptor: | Peptidyl-tRNA hydrolase, URIDINE | | Authors: | Kaushik, S, Singh, N, Yamini, S, Singh, A, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2013-03-28 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mode of Inhibitor Binding to Peptidyl-tRNA Hydrolase: Binding Studies and Structure Determination of Unbound and Bound Peptidyl-tRNA Hydrolase from Acinetobacter baumannii
Plos One, 8, 2013
|
|
1S0C
 
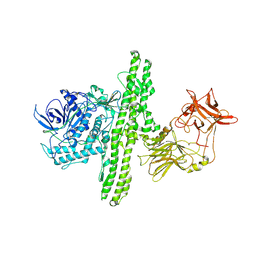 | | Crystal structure of botulinum neurotoxin type B at pH 5.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1XEV
 
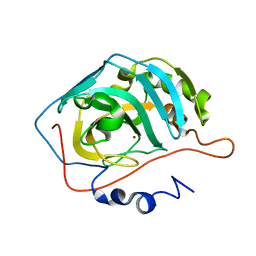 | |
1XEG
 
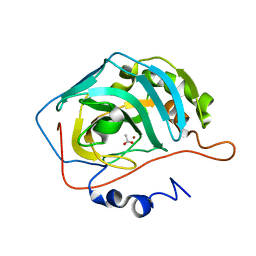 | | Crystal structure of human carbonic anhydrase II complexed with an acetate ion | | Descriptor: | ACETATE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Mazumdar, P.A, Kumaran, D, Das, A.K, Swaminathan, S. | | Deposit date: | 2004-09-10 | | Release date: | 2005-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A novel acetate-bound complex of human carbonic anhydrase II.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
3LOP
 
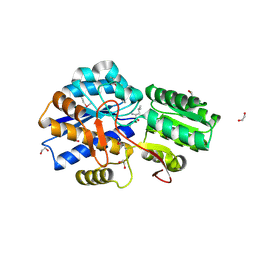 | | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum | | Descriptor: | 1,2-ETHANEDIOL, LEUCINE, MAGNESIUM ION, ... | | Authors: | Palani, K, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-04 | | Release date: | 2010-02-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of substrate-binding periplasmic protein (Pbp) from Ralstonia solanacearum
To be Published
|
|
1U02
 
 | | Crystal structure of trehalose-6-phosphate phosphatase related protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Krishnamurthy, N.R, Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-12 | | Release date: | 2004-07-20 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications.
Protein Sci., 15, 2006
|
|
2HAE
 
 | |
1TXN
 
 | |
3STP
 
 | | Crystal structure of a putative galactonate dehydratase | | Descriptor: | Galactonate dehydratase, putative, MAGNESIUM ION | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-07-11 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of a putative galactonate dehydratase
To be Published
|
|
1S0F
 
 | | Crystal structure of botulinum neurotoxin type B at pH 7.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
3TCS
 
 | | Crystal structure of a putative racemase from Roseobacter denitrificans | | Descriptor: | CHLORIDE ION, D-ALANINE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-09 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of a putative racemase from Roseobacter denitrificans
To be Published
|
|
1S0D
 
 | | Crystal structure of botulinum neurotoxin type B at pH 5.5 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
